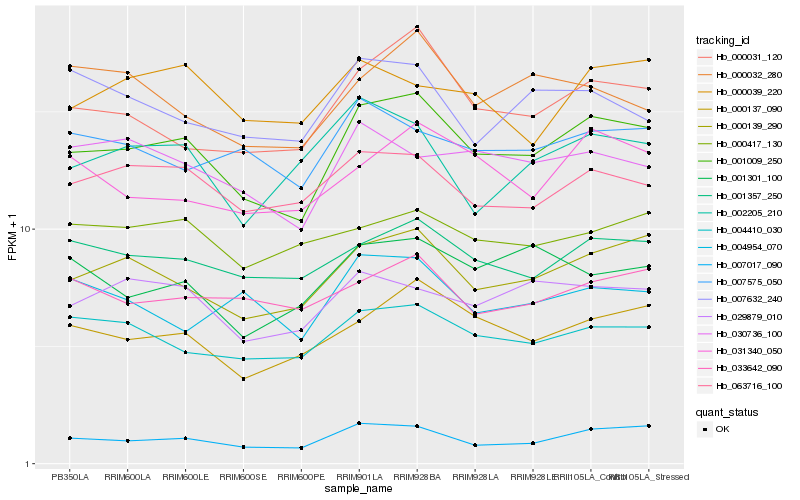
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001009_250 |
0.0 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007017_090 |
0.0654337315 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 186 [Jatropha curcas] |
| 3 |
Hb_000139_290 |
0.0689019734 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000137_090 |
0.0813318873 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Vitis vinifera] |
| 5 |
Hb_000031_120 |
0.0814576677 |
- |
- |
PREDICTED: uncharacterized protein LOC105637488 [Jatropha curcas] |
| 6 |
Hb_001357_250 |
0.0861728124 |
- |
- |
PREDICTED: UPF0505 protein C16orf62 homolog [Jatropha curcas] |
| 7 |
Hb_004410_030 |
0.0877515075 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_063716_100 |
0.0908428469 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_029879_010 |
0.0933856452 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 10 |
Hb_031340_050 |
0.0976481232 |
- |
- |
PREDICTED: tropinone reductase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_030736_100 |
0.1012418202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_033642_090 |
0.1015426482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001301_100 |
0.1016022032 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_007575_050 |
0.1018408034 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 15 |
Hb_000032_280 |
0.105406579 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 16 |
Hb_000417_130 |
0.106606863 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_002205_210 |
0.1070010509 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004954_070 |
0.1070770755 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 19 |
Hb_007632_240 |
0.1074650238 |
- |
- |
transporter, putative [Ricinus communis] |
| 20 |
Hb_000039_220 |
0.1081703931 |
- |
- |
PREDICTED: vesicle-associated protein 4-2-like [Gossypium raimondii] |