| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
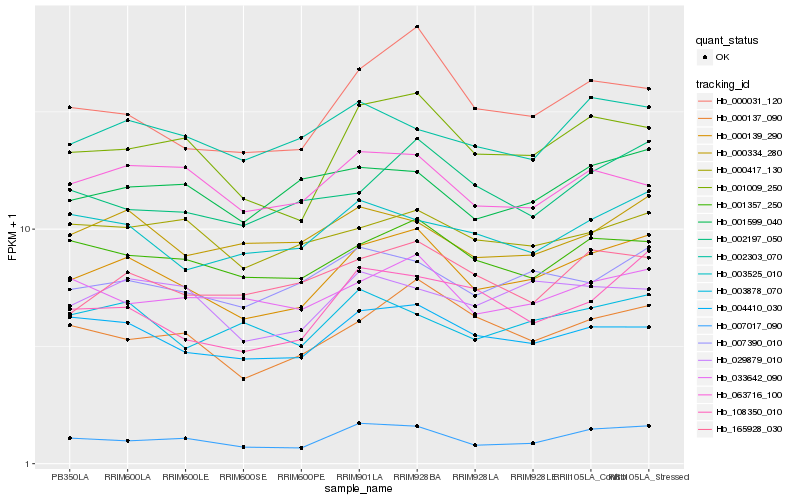
Hb_000139_290 |
0.0 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_007017_090 |
0.0673486022 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 186 [Jatropha curcas] |
| 3 |
Hb_001009_250 |
0.0689019734 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004410_030 |
0.0738212342 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001357_250 |
0.078702869 |
- |
- |
PREDICTED: UPF0505 protein C16orf62 homolog [Jatropha curcas] |
| 6 |
Hb_000137_090 |
0.0792994238 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Vitis vinifera] |
| 7 |
Hb_007390_010 |
0.0805023389 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_000031_120 |
0.0806549906 |
- |
- |
PREDICTED: uncharacterized protein LOC105637488 [Jatropha curcas] |
| 9 |
Hb_108350_010 |
0.0848183381 |
- |
- |
GTP-binding protein erg, putative [Ricinus communis] |
| 10 |
Hb_165928_030 |
0.0862216602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_033642_090 |
0.0865565625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002197_050 |
0.0869070935 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |
| 13 |
Hb_001599_040 |
0.0874525659 |
- |
- |
PREDICTED: suppressor protein SRP40 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003525_010 |
0.0879928323 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 24-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000334_280 |
0.0886755365 |
- |
- |
PREDICTED: protein FRA10AC1 [Jatropha curcas] |
| 16 |
Hb_000417_130 |
0.0899090657 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_029879_010 |
0.0903238887 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 18 |
Hb_063716_100 |
0.0912247608 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_003878_070 |
0.0921552853 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 20 |
Hb_002303_070 |
0.0924837357 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |