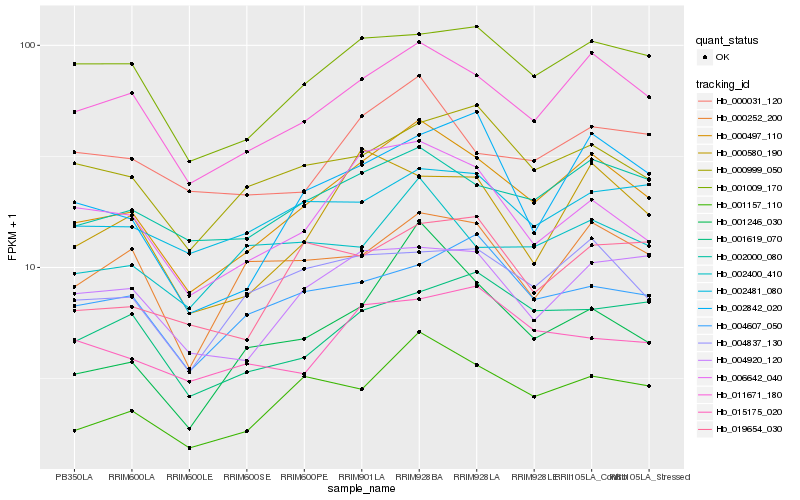
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 2 |
Hb_011671_180 |
0.06430601 |
- |
- |
PREDICTED: probable quinone oxidoreductase [Jatropha curcas] |
| 3 |
Hb_001157_110 |
0.0836226292 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 4 |
Hb_002000_080 |
0.0874012159 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006642_040 |
0.0920351416 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 6 |
Hb_001009_170 |
0.0954208599 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_004837_130 |
0.1004857014 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 8 |
Hb_004920_120 |
0.1019204971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000031_120 |
0.1029413066 |
- |
- |
PREDICTED: uncharacterized protein LOC105637488 [Jatropha curcas] |
| 10 |
Hb_000999_050 |
0.1043986245 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000580_190 |
0.1082090011 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000252_200 |
0.1086732866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002400_410 |
0.1100352887 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 14 |
Hb_002842_020 |
0.1110261942 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 15 |
Hb_002481_080 |
0.1139815456 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_019654_030 |
0.1148506571 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 17 |
Hb_001619_070 |
0.116644968 |
- |
- |
PREDICTED: uncharacterized protein LOC103503585 [Cucumis melo] |
| 18 |
Hb_001246_030 |
0.1169642521 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 19 |
Hb_004607_050 |
0.1174610755 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_015175_020 |
0.1176781616 |
- |
- |
hypothetical protein JCGZ_14452 [Jatropha curcas] |