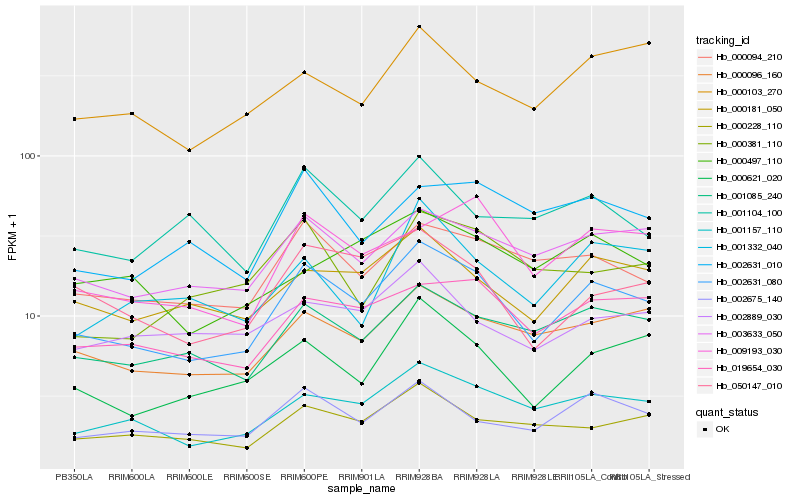
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_080 |
0.0 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001157_110 |
0.0941267956 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 3 |
Hb_001085_240 |
0.1068336275 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003633_050 |
0.1071544229 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 5 |
Hb_000096_160 |
0.1086929061 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 6 |
Hb_000094_210 |
0.1115933101 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 7 |
Hb_002675_140 |
0.1116512969 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 8 |
Hb_019654_030 |
0.1122915059 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_000621_020 |
0.1123372433 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 10 |
Hb_000181_050 |
0.1127271203 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 11 |
Hb_050147_010 |
0.1161278023 |
- |
- |
hypothetical protein B456_005G052100 [Gossypium raimondii] |
| 12 |
Hb_000228_110 |
0.1208476536 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_009193_030 |
0.1265998499 |
- |
- |
hypothetical protein POPTR_0010s16620g [Populus trichocarpa] |
| 14 |
Hb_002631_010 |
0.136716193 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 15 |
Hb_000103_270 |
0.1370293122 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_002889_030 |
0.1380582115 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 17 |
Hb_001104_100 |
0.1392176475 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 18 |
Hb_000381_110 |
0.1409433305 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000497_110 |
0.1421225609 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 20 |
Hb_001332_040 |
0.1423490289 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |