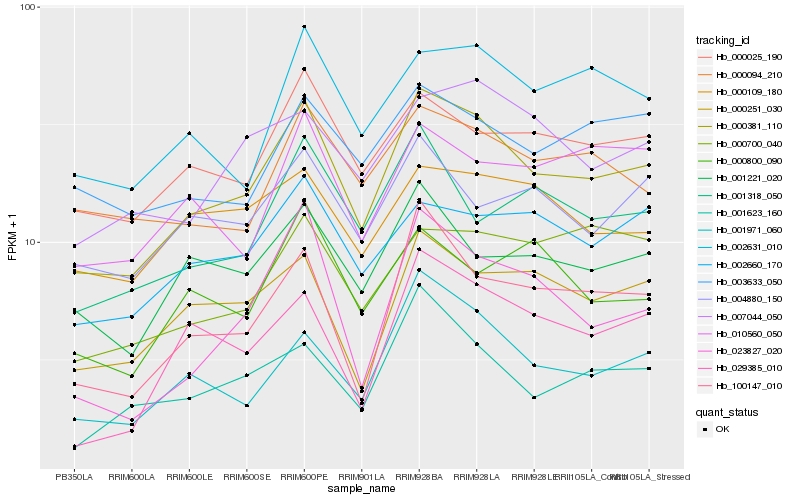
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000381_110 |
0.0 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001971_060 |
0.1013423861 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Populus euphratica] |
| 3 |
Hb_100147_010 |
0.1062569771 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 4 |
Hb_000025_190 |
0.1068779901 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 5 |
Hb_000251_030 |
0.1075180201 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001221_020 |
0.1096810613 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 7 |
Hb_002631_010 |
0.1118080284 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 8 |
Hb_002660_170 |
0.1121838364 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_001623_160 |
0.1144150396 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 10 |
Hb_001318_050 |
0.1167339589 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 11 |
Hb_000094_210 |
0.1185205796 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 12 |
Hb_007044_050 |
0.1185506629 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 13 |
Hb_004880_150 |
0.1207562608 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 14 |
Hb_029385_010 |
0.1221140718 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 15 |
Hb_010560_050 |
0.1222404784 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 16 |
Hb_003633_050 |
0.1237490301 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 17 |
Hb_023827_020 |
0.1243938667 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 18 |
Hb_000109_180 |
0.1246006042 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 19 |
Hb_000700_040 |
0.1261434182 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |
| 20 |
Hb_000800_090 |
0.1277483926 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |