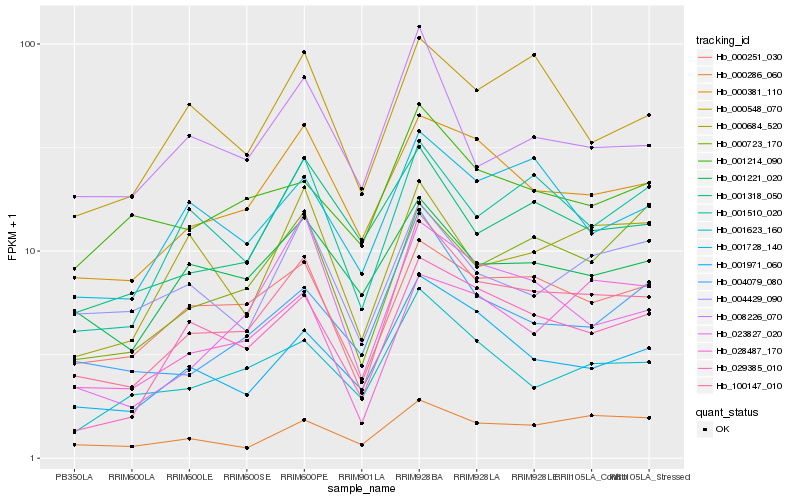
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100147_010 |
0.0 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 2 |
Hb_000251_030 |
0.1056457075 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000381_110 |
0.1062569771 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_029385_010 |
0.1102582314 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 5 |
Hb_001971_060 |
0.11155511 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Populus euphratica] |
| 6 |
Hb_001510_020 |
0.116072372 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 7 |
Hb_001623_160 |
0.1244519263 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 8 |
Hb_000286_060 |
0.1254972436 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 9 |
Hb_001728_140 |
0.1258572704 |
- |
- |
- |
| 10 |
Hb_004079_080 |
0.1262385308 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 11 |
Hb_000548_070 |
0.1324572311 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 12 |
Hb_008226_070 |
0.1345538542 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 13 |
Hb_023827_020 |
0.1347972404 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 14 |
Hb_001221_020 |
0.1354946634 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 15 |
Hb_001318_050 |
0.1357197178 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 16 |
Hb_000723_170 |
0.136442808 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 17 |
Hb_000684_520 |
0.1364810995 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 18 |
Hb_004429_090 |
0.1402879078 |
- |
- |
PREDICTED: ras GTPase-activating protein-binding protein 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001214_090 |
0.141935907 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 20 |
Hb_028487_170 |
0.1432125168 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |