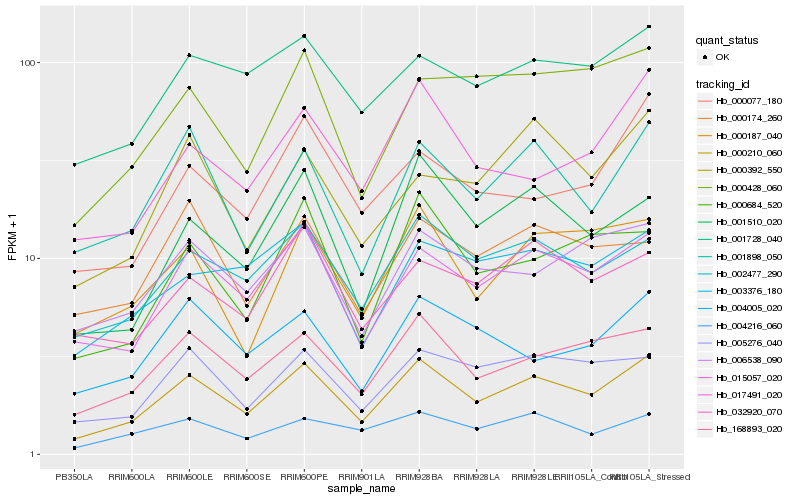
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_060 |
0.0 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 2 |
Hb_015057_020 |
0.082293825 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 3 |
Hb_168893_020 |
0.087489064 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 4 |
Hb_002477_290 |
0.0968489294 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001898_050 |
0.1008440437 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 6 |
Hb_001510_020 |
0.104013331 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 7 |
Hb_005276_040 |
0.1042793789 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 8 |
Hb_000187_040 |
0.1112062496 |
- |
- |
PREDICTED: pyridoxal kinase isoform X5 [Vitis vinifera] |
| 9 |
Hb_006538_090 |
0.1121807914 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 10 |
Hb_004216_060 |
0.1124267132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000684_520 |
0.1140222482 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 12 |
Hb_001728_040 |
0.1149278238 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 13 |
Hb_000428_060 |
0.1186922236 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_003376_180 |
0.1195552296 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 15 |
Hb_017491_020 |
0.1199263561 |
- |
- |
hypothetical protein B456_011G056700 [Gossypium raimondii] |
| 16 |
Hb_004005_020 |
0.1215423038 |
- |
- |
PREDICTED: nudix hydrolase 20, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_032920_070 |
0.1225536995 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 18 |
Hb_000077_180 |
0.1256064211 |
- |
- |
- |
| 19 |
Hb_000392_550 |
0.1260501746 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000174_260 |
0.1266725108 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |