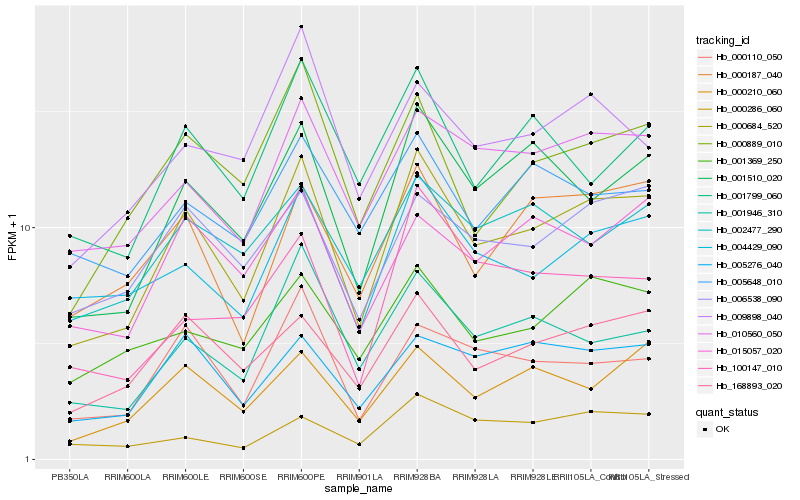
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_520 |
0.0 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 2 |
Hb_001510_020 |
0.0999520471 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 3 |
Hb_168893_020 |
0.1040880255 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 4 |
Hb_010560_050 |
0.1042687167 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 5 |
Hb_004429_090 |
0.1094350195 |
- |
- |
PREDICTED: ras GTPase-activating protein-binding protein 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000187_040 |
0.111727376 |
- |
- |
PREDICTED: pyridoxal kinase isoform X5 [Vitis vinifera] |
| 7 |
Hb_000210_060 |
0.1140222482 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 8 |
Hb_006538_090 |
0.1170993346 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 9 |
Hb_000889_010 |
0.1177878934 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 10 |
Hb_000110_050 |
0.1188003191 |
- |
- |
- |
| 11 |
Hb_005276_040 |
0.1203849487 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 12 |
Hb_001946_310 |
0.1226562634 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 13 |
Hb_001369_250 |
0.1226637445 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 14 |
Hb_015057_020 |
0.1247970238 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 15 |
Hb_002477_290 |
0.129175131 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_001799_060 |
0.1312043411 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 17 |
Hb_000286_060 |
0.1344674084 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 18 |
Hb_005648_010 |
0.13527526 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 19 |
Hb_009898_040 |
0.1355190949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_100147_010 |
0.1364810995 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |