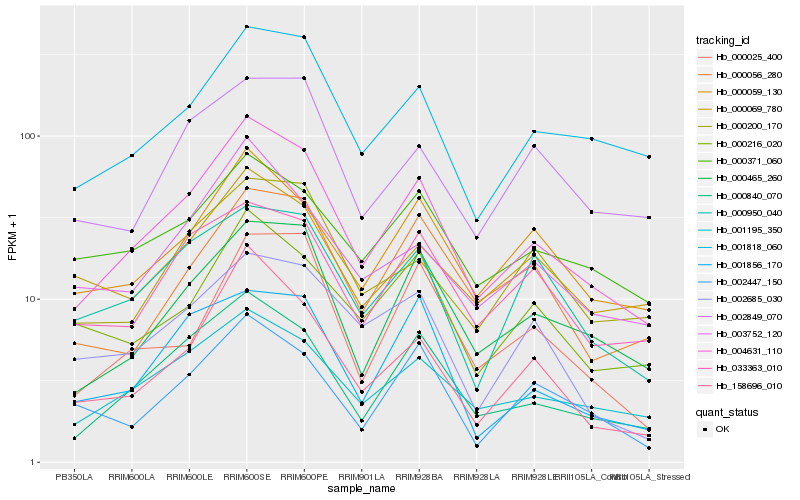
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_110 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_000840_070 |
0.0850170188 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_001195_350 |
0.1056679507 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 4 |
Hb_000465_260 |
0.1098839562 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 5 |
Hb_001818_060 |
0.1176719266 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 6 |
Hb_158696_010 |
0.1186218868 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 7 |
Hb_000059_130 |
0.1214912089 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 8 |
Hb_003752_120 |
0.1227605932 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 9 |
Hb_033363_010 |
0.1298826145 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 10 |
Hb_000216_020 |
0.1312677944 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 11 |
Hb_000056_280 |
0.1384811498 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 12 |
Hb_000200_170 |
0.1389326357 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 13 |
Hb_000371_060 |
0.1409477676 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 14 |
Hb_000950_040 |
0.1411225259 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 15 |
Hb_000025_400 |
0.1420103129 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 16 |
Hb_001856_170 |
0.1420896653 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 17 |
Hb_002849_070 |
0.1428719003 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000069_780 |
0.144036493 |
- |
- |
atpob1, putative [Ricinus communis] |
| 19 |
Hb_002447_150 |
0.1444540768 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_002685_030 |
0.1452413728 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |