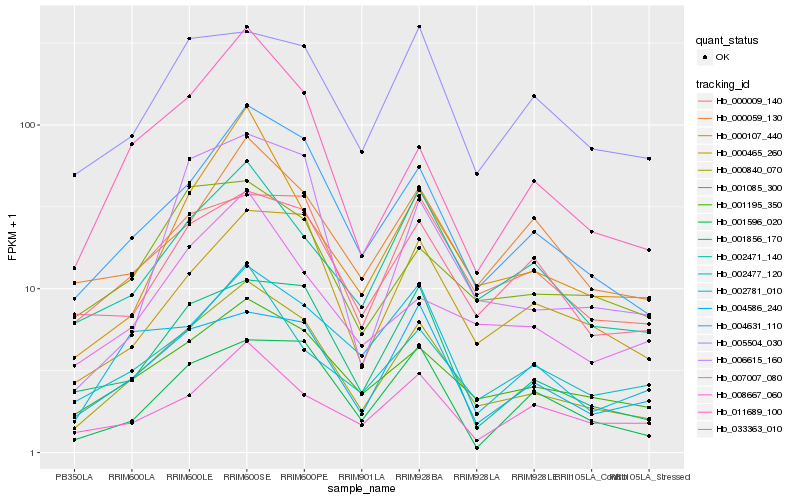
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000840_070 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_004631_110 |
0.0850170188 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_001195_350 |
0.1065003812 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 4 |
Hb_002471_140 |
0.1233819815 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 5 |
Hb_001856_170 |
0.1302556701 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 6 |
Hb_000465_260 |
0.1311527196 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 7 |
Hb_000009_140 |
0.1321867546 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 8 |
Hb_004586_240 |
0.1336364296 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 9 |
Hb_033363_010 |
0.1375425856 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 10 |
Hb_002781_010 |
0.1404548879 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_011689_100 |
0.1467799092 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 12 |
Hb_002477_120 |
0.1469658345 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_005504_030 |
0.1496580019 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 14 |
Hb_000059_130 |
0.1505909824 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 15 |
Hb_001085_300 |
0.153274844 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 16 |
Hb_008667_060 |
0.1537196533 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 17 |
Hb_000107_440 |
0.1544189404 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 18 |
Hb_007007_080 |
0.1575405313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001596_020 |
0.1581934753 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 20 |
Hb_006615_160 |
0.1589624394 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |