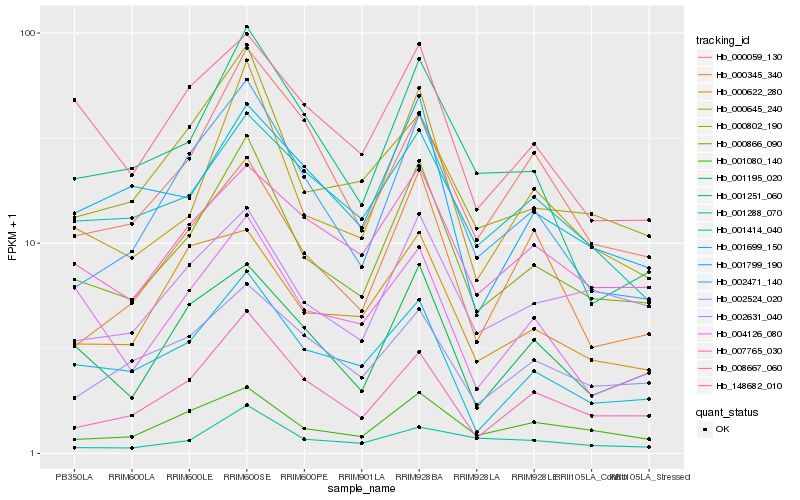
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_090 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_000645_240 |
0.0844697542 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 3 |
Hb_001080_140 |
0.1001714007 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002471_140 |
0.1079325467 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 5 |
Hb_008667_060 |
0.1099136578 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 6 |
Hb_000802_190 |
0.1100570177 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 7 |
Hb_001699_150 |
0.1112943118 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 8 |
Hb_002631_040 |
0.1128704182 |
- |
- |
hypothetical protein EUGRSUZ_J03178 [Eucalyptus grandis] |
| 9 |
Hb_001195_020 |
0.1152401391 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007765_030 |
0.1170410388 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 11 |
Hb_000345_340 |
0.1194749136 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 12 |
Hb_002524_020 |
0.1289990089 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 13 |
Hb_001414_040 |
0.1290507314 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001251_060 |
0.1296253558 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 15 |
Hb_001799_190 |
0.1298796618 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 16 |
Hb_148682_010 |
0.1307681766 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 17 |
Hb_004126_080 |
0.1310884947 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 18 |
Hb_001288_070 |
0.1324684852 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 19 |
Hb_000059_130 |
0.1332206119 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000622_280 |
0.1336998129 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |