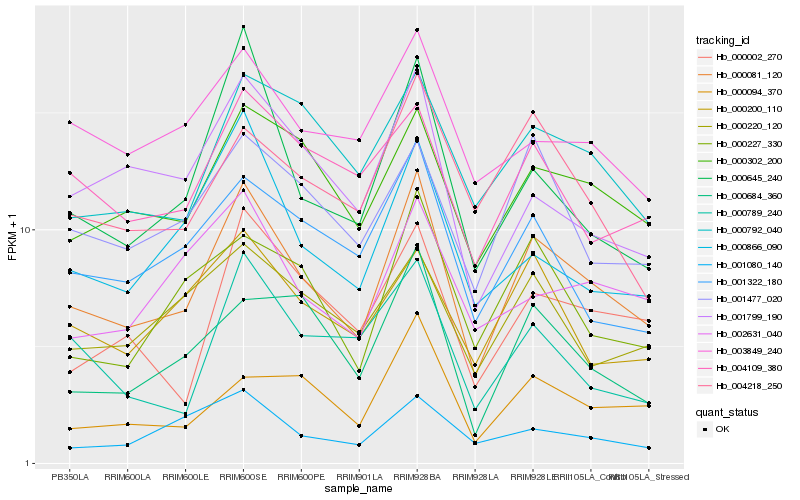
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000081_120 |
0.0 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 2 |
Hb_000302_200 |
0.1263052504 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 3 |
Hb_000002_270 |
0.1383558387 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 4 |
Hb_000792_040 |
0.1384756374 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 5 |
Hb_000645_240 |
0.138889481 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 6 |
Hb_001322_180 |
0.1392884275 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 7 |
Hb_000227_330 |
0.1394005179 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 8 |
Hb_001477_020 |
0.140741624 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000866_090 |
0.1436820395 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 10 |
Hb_000684_360 |
0.1445861724 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 11 |
Hb_004218_250 |
0.1454771817 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 12 |
Hb_000094_370 |
0.146238848 |
- |
- |
- |
| 13 |
Hb_001799_190 |
0.1466285376 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 14 |
Hb_000200_110 |
0.1476131998 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002631_040 |
0.1487732146 |
- |
- |
hypothetical protein EUGRSUZ_J03178 [Eucalyptus grandis] |
| 16 |
Hb_004109_380 |
0.1492168289 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 17 |
Hb_000789_240 |
0.1496838106 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 18 |
Hb_000220_120 |
0.1497162482 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003849_240 |
0.1498461691 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001080_140 |
0.1503939032 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |