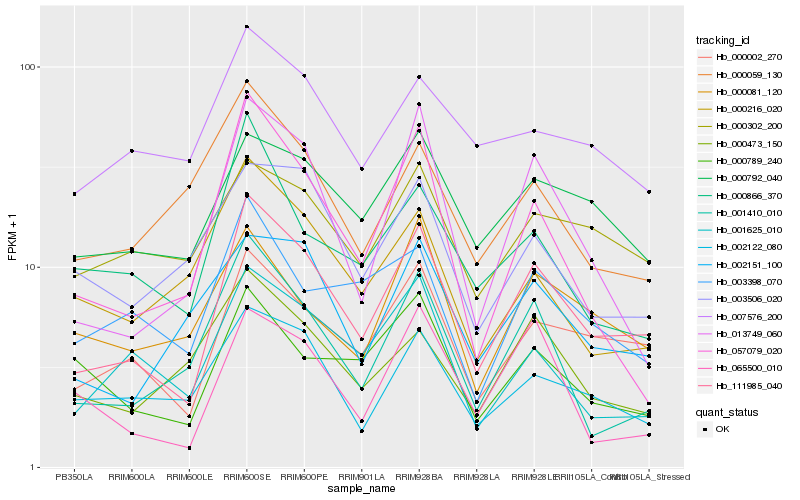
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111985_040 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 2 |
Hb_000002_270 |
0.1073535989 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 3 |
Hb_013749_060 |
0.1480506594 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 4 |
Hb_001410_010 |
0.1512608782 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 5 |
Hb_000792_040 |
0.1565860203 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 6 |
Hb_002122_080 |
0.1570482205 |
- |
- |
- |
| 7 |
Hb_001625_010 |
0.1591672826 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 8 |
Hb_057079_020 |
0.1628429948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002151_100 |
0.1637579954 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 10 |
Hb_003398_070 |
0.1662797774 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 11 |
Hb_000789_240 |
0.168459999 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 12 |
Hb_000866_370 |
0.1686030585 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 13 |
Hb_065500_010 |
0.1688563749 |
- |
- |
- |
| 14 |
Hb_000081_120 |
0.1690729066 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 15 |
Hb_007576_200 |
0.1711482493 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_000473_150 |
0.1716340048 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000302_200 |
0.1729602155 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 18 |
Hb_003506_020 |
0.1758158056 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 19 |
Hb_000059_130 |
0.1759502103 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000216_020 |
0.1766267723 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |