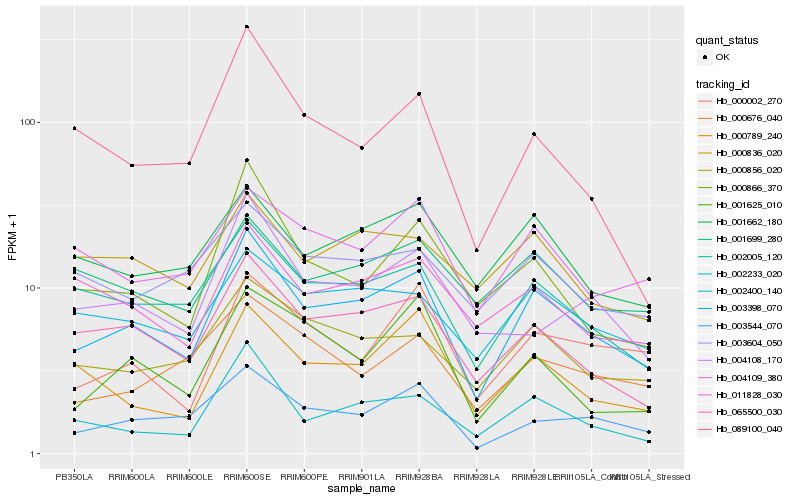
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003398_070 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 2 |
Hb_065500_030 |
0.1130058995 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 3 |
Hb_002005_120 |
0.119597215 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 4 |
Hb_002233_020 |
0.1219727601 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 5 |
Hb_003544_070 |
0.1239339133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000856_020 |
0.1329493079 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 7 |
Hb_002400_140 |
0.1343810744 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 8 |
Hb_001662_180 |
0.1395880809 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 9 |
Hb_000866_370 |
0.1401265172 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 10 |
Hb_000789_240 |
0.1420738971 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 11 |
Hb_000002_270 |
0.1434040762 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 12 |
Hb_004109_380 |
0.1450690302 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 13 |
Hb_011828_030 |
0.145545078 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 14 |
Hb_004108_170 |
0.1513899188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000676_040 |
0.1519247033 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 16 |
Hb_001625_010 |
0.1527611911 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 17 |
Hb_001699_280 |
0.1536613809 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 18 |
Hb_000836_020 |
0.1539771492 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 19 |
Hb_003604_050 |
0.1557321676 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_089100_040 |
0.1574236025 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |