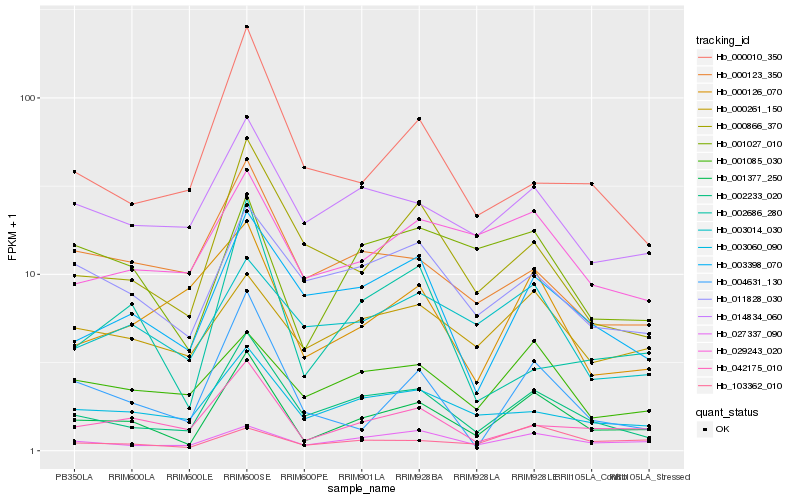
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_250 |
0.0 |
- |
- |
dynamin, putative [Ricinus communis] |
| 2 |
Hb_002233_020 |
0.1479100603 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 3 |
Hb_042175_010 |
0.1688175987 |
- |
- |
hypothetical protein CISIN_1g0016721mg, partial [Citrus sinensis] |
| 4 |
Hb_027337_090 |
0.1875092853 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 5 |
Hb_000866_370 |
0.1904005588 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 6 |
Hb_004631_130 |
0.1916079361 |
- |
- |
helicase domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_003398_070 |
0.192290788 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 8 |
Hb_003060_090 |
0.1959529533 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 9 |
Hb_001027_010 |
0.1970360893 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0019s03720g [Populus trichocarpa] |
| 10 |
Hb_014834_060 |
0.2035347928 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001085_030 |
0.2044663046 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 12 |
Hb_103362_010 |
0.2078391057 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_011828_030 |
0.2097055174 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 14 |
Hb_002686_280 |
0.2129416328 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_029243_020 |
0.213302406 |
- |
- |
ENTH/VHS/GAT family protein [Theobroma cacao] |
| 16 |
Hb_000261_150 |
0.2137748796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000126_070 |
0.2145336856 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |
| 18 |
Hb_000123_350 |
0.2159094682 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000010_350 |
0.2172683577 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 20 |
Hb_003014_030 |
0.2188877657 |
- |
- |
PREDICTED: thyroid adenoma-associated protein homolog [Jatropha curcas] |