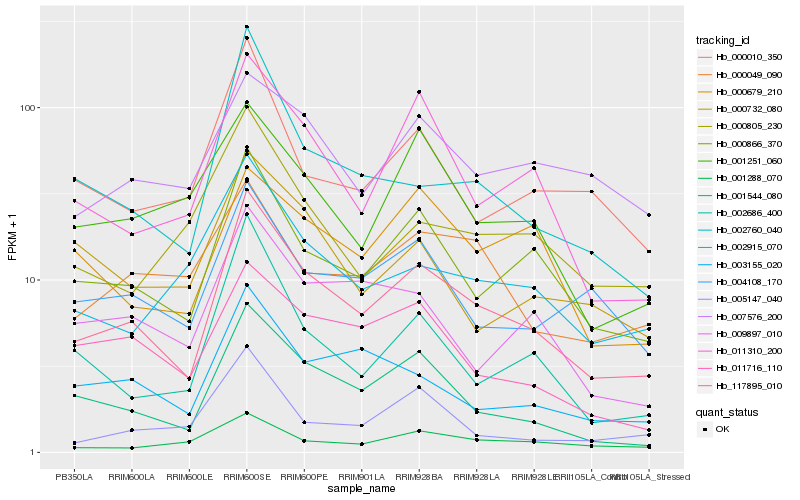
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_117895_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 2 |
Hb_000866_370 |
0.1111753991 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 3 |
Hb_001544_080 |
0.1231981448 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 4 |
Hb_011310_200 |
0.1438276014 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 5 |
Hb_004108_170 |
0.1545936383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002760_040 |
0.1553751629 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 7 |
Hb_003155_020 |
0.156982955 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 8 |
Hb_002915_070 |
0.1596978794 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 9 |
Hb_011716_110 |
0.1599348689 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 10 |
Hb_000010_350 |
0.1602026791 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 11 |
Hb_002686_400 |
0.1603197846 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 12 |
Hb_009897_010 |
0.1614698521 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_001288_070 |
0.1647622586 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 14 |
Hb_001251_060 |
0.1710937763 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 15 |
Hb_000805_230 |
0.1713997729 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 16 |
Hb_005147_040 |
0.1726116382 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 17 |
Hb_007576_200 |
0.1727867951 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_000049_090 |
0.1732361501 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000679_210 |
0.1756753835 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 20 |
Hb_000732_080 |
0.1757877499 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |