| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
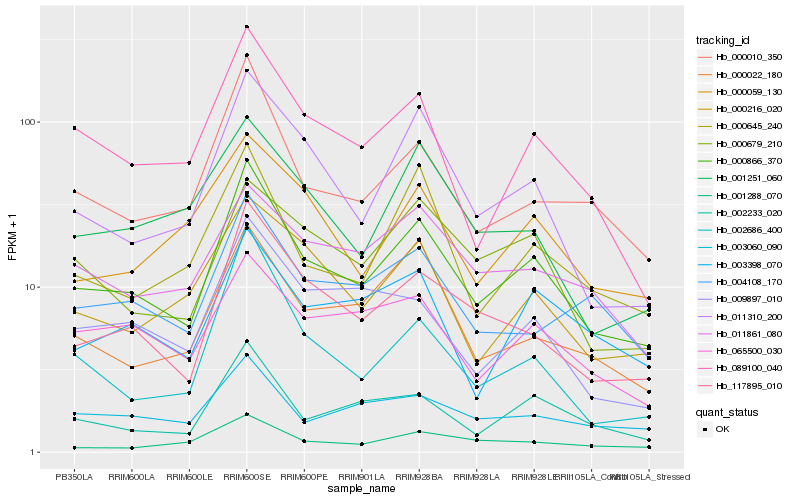
Hb_000866_370 |
0.0 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 2 |
Hb_117895_010 |
0.1111753991 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 3 |
Hb_000010_350 |
0.1144485703 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 4 |
Hb_089100_040 |
0.1197376608 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 5 |
Hb_002233_020 |
0.1210781728 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 6 |
Hb_011310_200 |
0.1217107014 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 7 |
Hb_000645_240 |
0.1356338162 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 8 |
Hb_065500_030 |
0.1372783327 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 9 |
Hb_000022_180 |
0.1374682462 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004108_170 |
0.1383960306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000216_020 |
0.1384990149 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 12 |
Hb_003398_070 |
0.1401265172 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 13 |
Hb_000679_210 |
0.1412519445 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 14 |
Hb_009897_010 |
0.1421947551 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_003060_090 |
0.1439922601 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 16 |
Hb_001288_070 |
0.1445034877 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 17 |
Hb_002686_400 |
0.1468362813 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 18 |
Hb_000059_130 |
0.1468921346 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 19 |
Hb_011861_080 |
0.1476492595 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 20 |
Hb_001251_060 |
0.1503222452 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |