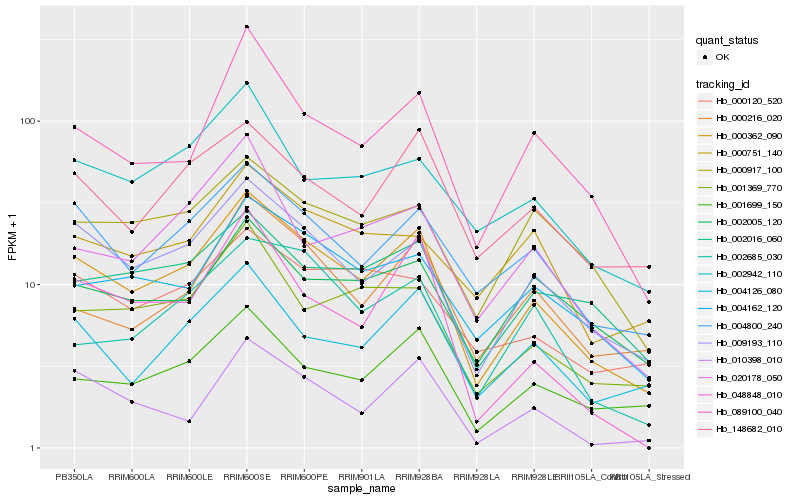
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009193_110 |
0.0897911453 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004800_240 |
0.1138055609 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 4 |
Hb_001699_150 |
0.118837657 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 5 |
Hb_089100_040 |
0.121250155 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 6 |
Hb_004162_120 |
0.1232553019 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 7 |
Hb_148682_010 |
0.1276017674 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 8 |
Hb_004126_080 |
0.1285210036 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 9 |
Hb_000216_020 |
0.1297185268 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 10 |
Hb_001369_770 |
0.1297658721 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 11 |
Hb_000120_520 |
0.1370355809 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 12 |
Hb_002016_060 |
0.1374154451 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 13 |
Hb_002942_110 |
0.1386729174 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 14 |
Hb_000917_100 |
0.142500416 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 15 |
Hb_002685_030 |
0.1434891692 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 16 |
Hb_020178_050 |
0.1437916972 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 17 |
Hb_010398_010 |
0.145241649 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 18 |
Hb_002005_120 |
0.1452800901 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 19 |
Hb_048848_010 |
0.1477870421 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 20 |
Hb_000751_140 |
0.1478218967 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |