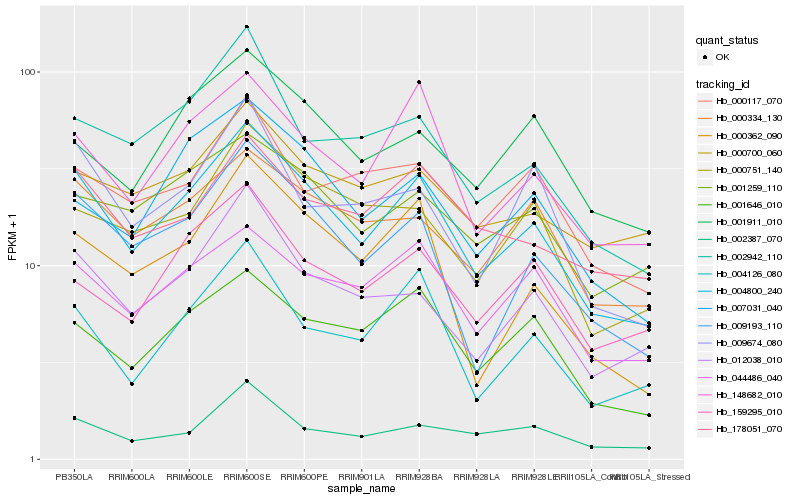
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_240 |
0.0 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 2 |
Hb_009193_110 |
0.0886748117 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_148682_010 |
0.0989726371 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 4 |
Hb_012038_010 |
0.1065394632 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 5 |
Hb_001646_010 |
0.1084051342 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 6 |
Hb_000334_130 |
0.1086178219 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 7 |
Hb_004126_080 |
0.1095212368 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 8 |
Hb_044486_040 |
0.1103420919 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 9 |
Hb_000362_090 |
0.1138055609 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007031_040 |
0.1145564622 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 11 |
Hb_001259_110 |
0.1156221547 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 12 |
Hb_002942_110 |
0.1158706184 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 13 |
Hb_178051_070 |
0.1193741067 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 14 |
Hb_002387_070 |
0.120192954 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g77010, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000751_140 |
0.1212748613 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 16 |
Hb_159295_010 |
0.1220278078 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 17 |
Hb_009674_080 |
0.1228028021 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 18 |
Hb_001911_010 |
0.1229585005 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 19 |
Hb_000700_060 |
0.1251857441 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 20 |
Hb_000117_070 |
0.1275705739 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |