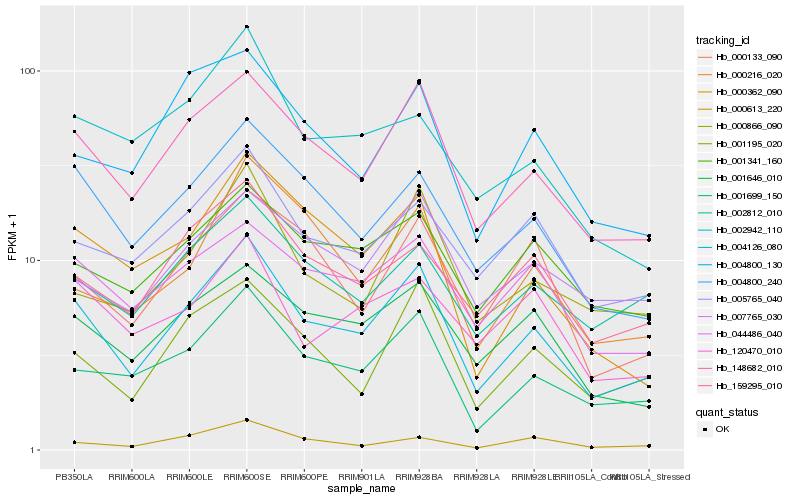
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004126_080 |
0.0 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 2 |
Hb_148682_010 |
0.0744084493 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 3 |
Hb_001699_150 |
0.1011186164 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 4 |
Hb_004800_240 |
0.1095212368 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 5 |
Hb_001646_010 |
0.117736579 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 6 |
Hb_001195_020 |
0.1189658292 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_159295_010 |
0.1249481116 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 8 |
Hb_005765_040 |
0.1256160289 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 9 |
Hb_044486_040 |
0.1268250518 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 10 |
Hb_002812_010 |
0.1283762814 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001341_160 |
0.1284804635 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000362_090 |
0.1285210036 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000866_090 |
0.1310884947 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_000133_090 |
0.1319840414 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 15 |
Hb_004800_130 |
0.1334217015 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 16 |
Hb_007765_030 |
0.1360003524 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 17 |
Hb_120470_010 |
0.1396570062 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 18 |
Hb_000216_020 |
0.1412330267 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 19 |
Hb_000613_220 |
0.141816016 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 20 |
Hb_002942_110 |
0.1441664523 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |