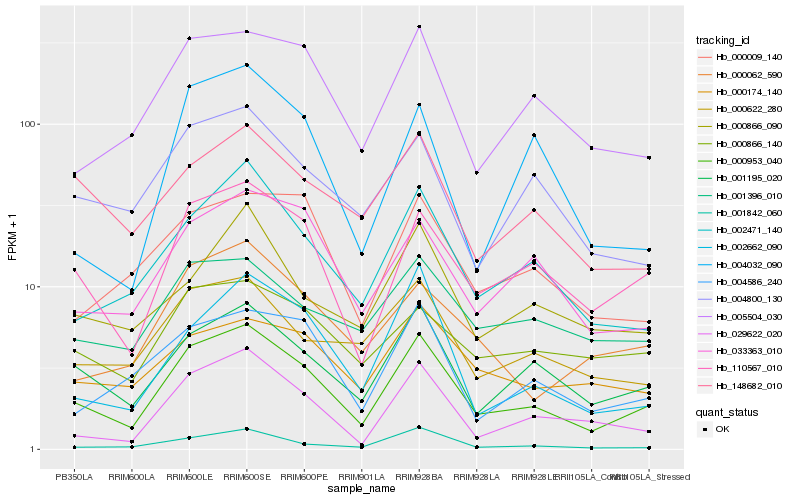
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000953_040 |
0.0 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 2 |
Hb_110567_010 |
0.1098577069 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001195_020 |
0.1146308201 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002471_140 |
0.1299346193 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 5 |
Hb_029622_020 |
0.1410731862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001842_060 |
0.1451368653 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 7 |
Hb_005504_030 |
0.1454000595 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 8 |
Hb_033363_010 |
0.1476728564 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 9 |
Hb_002662_090 |
0.1495672959 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 10 |
Hb_004586_240 |
0.1507159254 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 11 |
Hb_000622_280 |
0.1569992553 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004800_130 |
0.1590882682 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 13 |
Hb_000062_590 |
0.159761565 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_000009_140 |
0.1613894914 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 15 |
Hb_000866_090 |
0.1630240365 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_000174_140 |
0.1640225812 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 17 |
Hb_000866_140 |
0.1648137804 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 18 |
Hb_148682_010 |
0.1653938766 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 19 |
Hb_004032_090 |
0.1654649836 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 20 |
Hb_001396_010 |
0.1663352009 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |