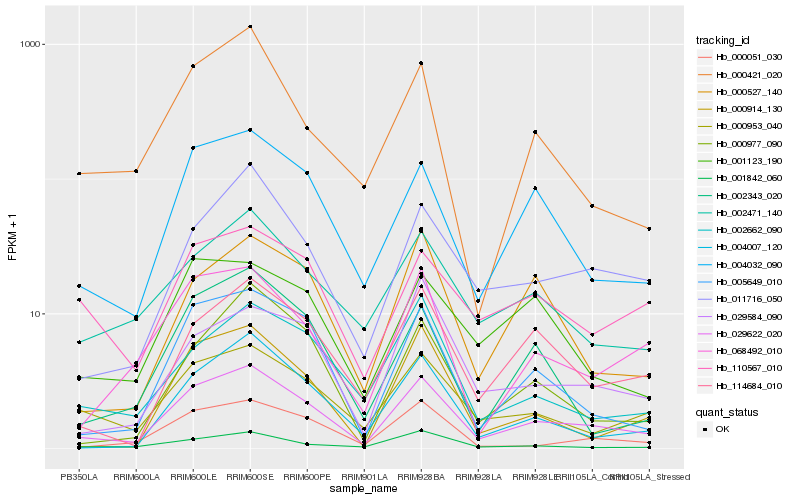
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029622_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004032_090 |
0.1317690988 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 3 |
Hb_011716_050 |
0.1389018386 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 4 |
Hb_005649_010 |
0.1407932341 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 5 |
Hb_000953_040 |
0.1410731862 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 6 |
Hb_114684_010 |
0.149892849 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 7 |
Hb_000051_030 |
0.1526999226 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 8 |
Hb_004007_120 |
0.1550801066 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 9 |
Hb_002471_140 |
0.1559843016 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 10 |
Hb_068492_010 |
0.156461042 |
- |
- |
- |
| 11 |
Hb_001842_060 |
0.1585821294 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 12 |
Hb_002662_090 |
0.1661119142 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 13 |
Hb_000977_090 |
0.1667786018 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 14 |
Hb_000421_020 |
0.1673404484 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 15 |
Hb_000527_140 |
0.1697268571 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 16 |
Hb_029584_090 |
0.1700902489 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 17 |
Hb_110567_010 |
0.1702090698 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002343_020 |
0.1712683135 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 19 |
Hb_000914_130 |
0.171489911 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 20 |
Hb_001123_190 |
0.1720974908 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |