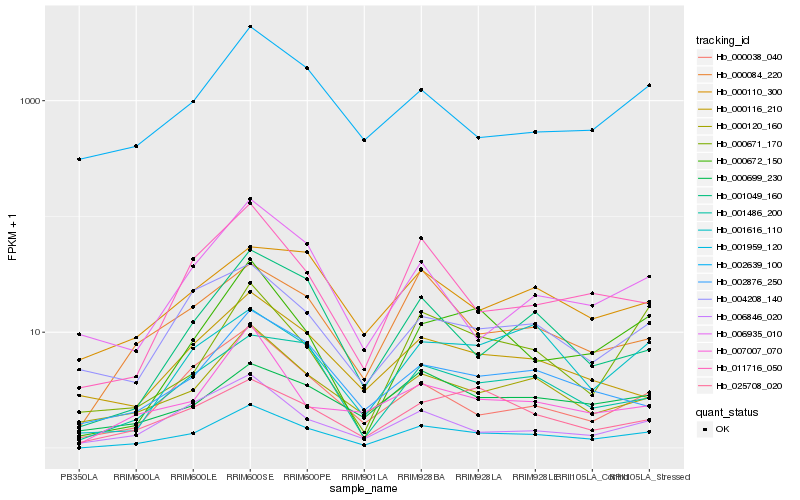
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 2 |
Hb_000120_160 |
0.1489236863 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_001486_200 |
0.1529164529 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 4 |
Hb_000672_150 |
0.1559061049 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 5 |
Hb_000084_220 |
0.1571871509 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 6 |
Hb_011716_050 |
0.1625954056 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 7 |
Hb_002876_250 |
0.1630356715 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 8 |
Hb_001049_160 |
0.1669063888 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 9 |
Hb_000038_040 |
0.1698223833 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 10 |
Hb_006846_020 |
0.1701391981 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 11 |
Hb_001616_110 |
0.1705683007 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 12 |
Hb_000699_230 |
0.1716741244 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 13 |
Hb_004208_140 |
0.1737065379 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 14 |
Hb_025708_020 |
0.1745315859 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_002639_100 |
0.1761224612 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 16 |
Hb_007007_070 |
0.1781481709 |
transcription factor |
TF Family: TCP |
hypothetical protein MIMGU_mgv1a022855mg, partial [Erythranthe guttata] |
| 17 |
Hb_000116_210 |
0.1808985559 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 18 |
Hb_000110_300 |
0.1830879171 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 19 |
Hb_006935_010 |
0.1831384985 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_000671_170 |
0.1837637364 |
- |
- |
kinase, putative [Ricinus communis] |