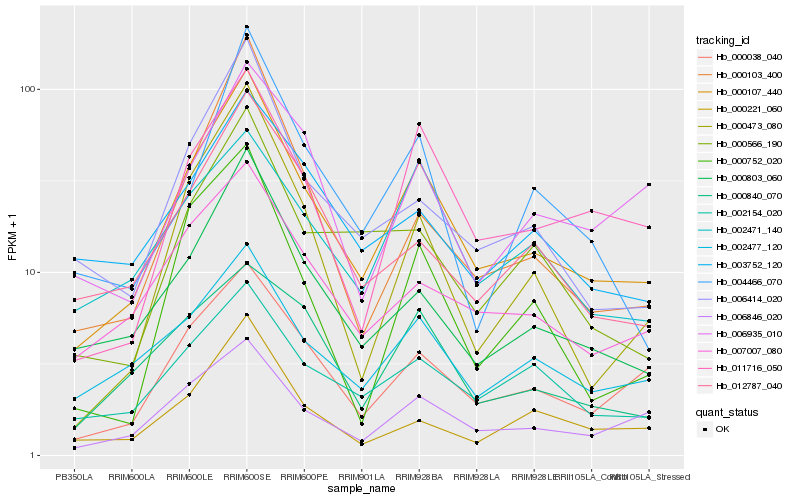
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_440 |
0.0 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 2 |
Hb_000038_040 |
0.1201489996 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 3 |
Hb_000803_060 |
0.1223220079 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 4 |
Hb_006846_020 |
0.1257835136 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 5 |
Hb_011716_050 |
0.1307873248 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 6 |
Hb_002477_120 |
0.1330404443 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_007007_080 |
0.1342714671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006414_020 |
0.136987865 |
transcription factor |
TF Family: MYB |
- |
| 9 |
Hb_002154_020 |
0.1415089974 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_012787_040 |
0.1450235886 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000566_190 |
0.1478659122 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000473_080 |
0.1492782076 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 13 |
Hb_000840_070 |
0.1544189404 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_002471_140 |
0.1552511227 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 15 |
Hb_003752_120 |
0.1565410573 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000221_060 |
0.1569699777 |
- |
- |
- |
| 17 |
Hb_004466_070 |
0.1593175679 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 18 |
Hb_006935_010 |
0.159804729 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 19 |
Hb_000752_020 |
0.1601101134 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000103_400 |
0.160310067 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |