| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
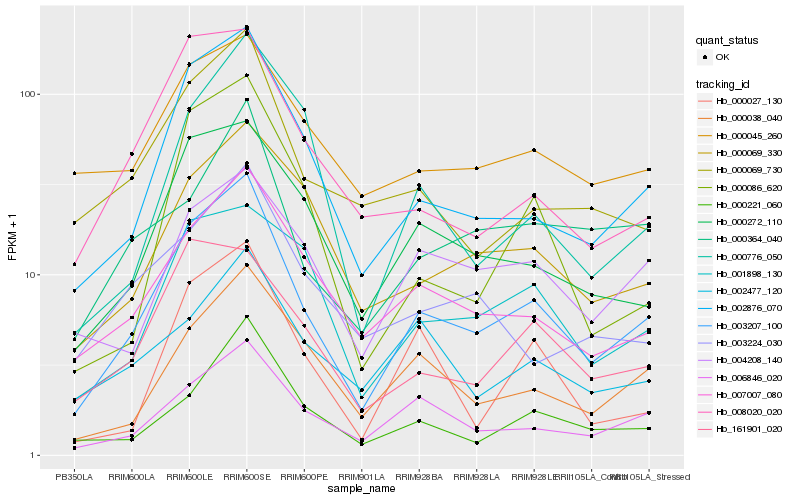
Hb_003207_100 |
0.0 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 2 |
Hb_002876_070 |
0.1152283968 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 3 |
Hb_006846_020 |
0.1373936727 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 4 |
Hb_007007_080 |
0.1415426458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000272_110 |
0.1482416512 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000038_040 |
0.1533627823 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 7 |
Hb_000069_330 |
0.1585645071 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000221_060 |
0.1606060574 |
- |
- |
- |
| 9 |
Hb_000069_730 |
0.1610231127 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_161901_020 |
0.1653068976 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 11 |
Hb_001898_130 |
0.1679849643 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000364_040 |
0.1708857435 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 13 |
Hb_000086_620 |
0.1721320907 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_008020_020 |
0.1722640875 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000045_260 |
0.1738263761 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_004208_140 |
0.1740633834 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 17 |
Hb_000027_130 |
0.1755437896 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 18 |
Hb_003224_030 |
0.17776895 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 19 |
Hb_002477_120 |
0.1790539454 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_000776_050 |
0.1799226892 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |