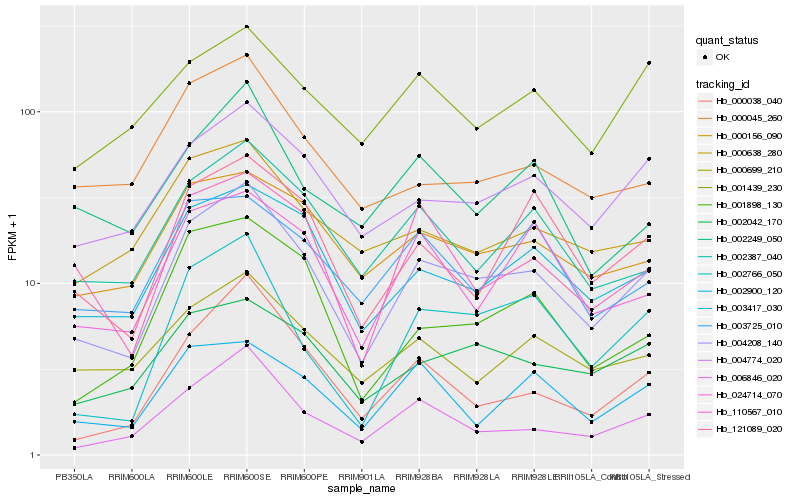
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004208_140 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 2 |
Hb_002900_120 |
0.1087955655 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 3 |
Hb_004774_020 |
0.1140538503 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 4 |
Hb_002387_040 |
0.1222858627 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 5 |
Hb_002042_170 |
0.1241558105 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 6 |
Hb_000156_090 |
0.1274257149 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002766_050 |
0.1289085877 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 8 |
Hb_006846_020 |
0.1298466792 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 9 |
Hb_000699_210 |
0.1311616949 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 10 |
Hb_121089_020 |
0.1321182985 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 11 |
Hb_110567_010 |
0.1359765934 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_024714_070 |
0.1378629999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 13 |
Hb_000038_040 |
0.1382948519 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 14 |
Hb_000638_280 |
0.13937273 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 15 |
Hb_003417_030 |
0.1413576741 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001898_130 |
0.1415050806 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000045_260 |
0.1436471618 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_001439_230 |
0.1458518452 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002249_050 |
0.1470336025 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 20 |
Hb_003725_010 |
0.147970808 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |