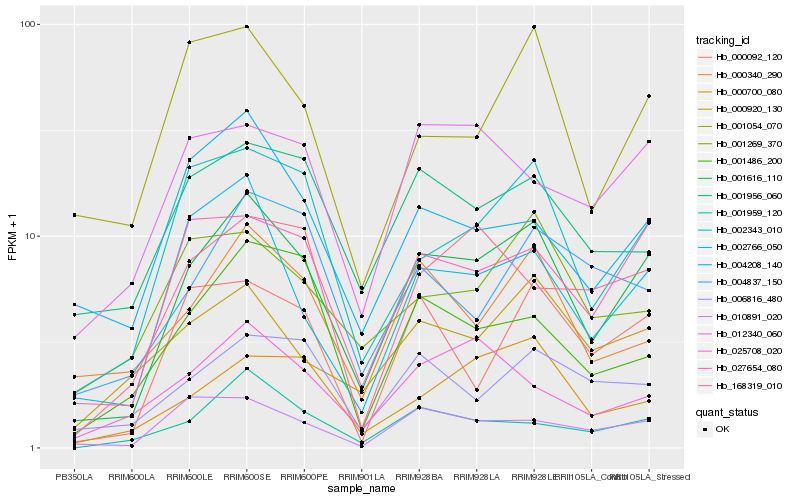
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001616_110 |
0.0 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 2 |
Hb_027654_080 |
0.1360574892 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002343_010 |
0.1458360586 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_002766_050 |
0.1579926482 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 5 |
Hb_000340_290 |
0.1660748916 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 6 |
Hb_000920_130 |
0.169142386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001959_120 |
0.1705683007 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 8 |
Hb_000700_080 |
0.1716943237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010891_020 |
0.1731414389 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 10 |
Hb_001054_070 |
0.1734604871 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 11 |
Hb_001956_060 |
0.1764185634 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 12 |
Hb_006816_480 |
0.1782134715 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012340_060 |
0.179376712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004208_140 |
0.1811929095 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 15 |
Hb_000092_120 |
0.1818643231 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 16 |
Hb_001269_370 |
0.1843492676 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 17 |
Hb_025708_020 |
0.188278984 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001486_200 |
0.1884906272 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 19 |
Hb_004837_150 |
0.1897865546 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 20 |
Hb_168319_010 |
0.1898430536 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |