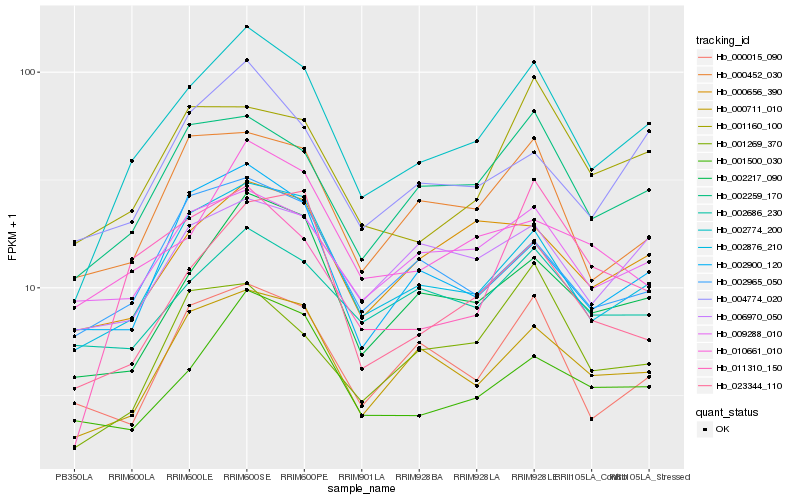
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002774_200 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 2 |
Hb_002876_210 |
0.0958251534 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 3 |
Hb_002259_170 |
0.1098072628 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_002217_090 |
0.1105139239 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 5 |
Hb_000711_010 |
0.1130497962 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 6 |
Hb_004774_020 |
0.1265856685 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 7 |
Hb_011310_150 |
0.1269366876 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 8 |
Hb_000452_030 |
0.1278750226 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 9 |
Hb_002965_050 |
0.1290753239 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 10 |
Hb_002900_120 |
0.1308093391 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 11 |
Hb_001500_030 |
0.1308449399 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000656_390 |
0.1309141412 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009288_010 |
0.1329194968 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001160_100 |
0.1354064316 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_023344_110 |
0.1362897551 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 16 |
Hb_001269_370 |
0.1377361411 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 17 |
Hb_006970_050 |
0.1383903131 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 18 |
Hb_000015_090 |
0.138844075 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 19 |
Hb_002686_230 |
0.139833005 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_010661_010 |
0.1414943562 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |