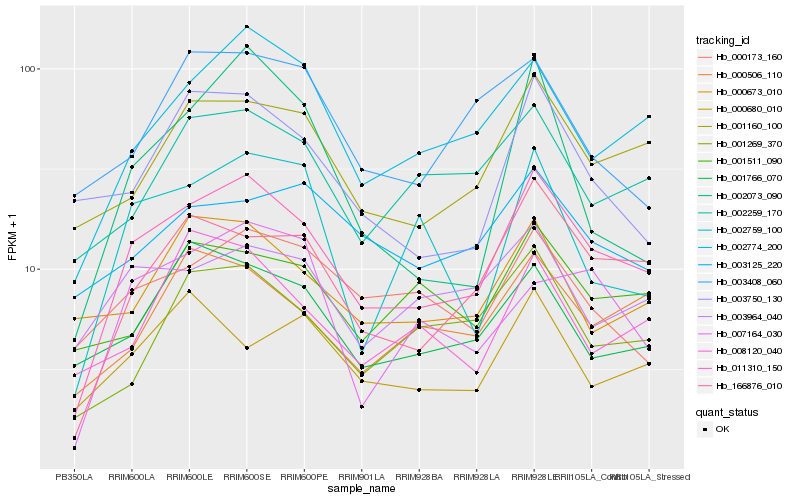
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_150 |
0.0 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 2 |
Hb_166876_010 |
0.1207584584 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 3 |
Hb_002774_200 |
0.1269366876 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 4 |
Hb_001160_100 |
0.1345672182 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002259_170 |
0.1530003552 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_001269_370 |
0.1538566628 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 7 |
Hb_000506_110 |
0.1566019958 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 8 |
Hb_001766_070 |
0.1623523561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003750_130 |
0.1624318306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003964_040 |
0.1629079341 |
- |
- |
PREDICTED: uncharacterized protein LOC105630528 [Jatropha curcas] |
| 11 |
Hb_000173_160 |
0.1647046311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000673_010 |
0.165557099 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001511_090 |
0.1677829974 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 14 |
Hb_002759_100 |
0.1696521355 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 15 |
Hb_008120_040 |
0.1724946382 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 16 |
Hb_003125_220 |
0.1742509704 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 17 |
Hb_003408_060 |
0.1768166793 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 18 |
Hb_000680_010 |
0.1772474487 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 19 |
Hb_007164_030 |
0.1774338145 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 20 |
Hb_002073_090 |
0.178536343 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |