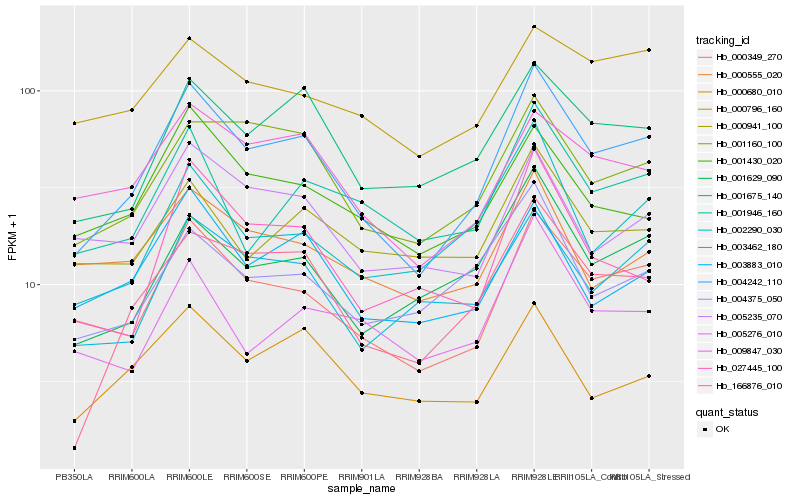
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004242_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 2 |
Hb_000349_270 |
0.0857855289 |
- |
- |
PREDICTED: uncharacterized protein ycf49 isoform X1 [Jatropha curcas] |
| 3 |
Hb_166876_010 |
0.1077485146 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 4 |
Hb_001675_140 |
0.1127925458 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 5 |
Hb_001160_100 |
0.1151286475 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001629_090 |
0.1166288646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009847_030 |
0.1236142097 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 8 |
Hb_003462_180 |
0.1249403848 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 9 |
Hb_003883_010 |
0.1254527667 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 10 |
Hb_002290_030 |
0.1269633507 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000680_010 |
0.1313109157 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 12 |
Hb_001946_160 |
0.1315695036 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 13 |
Hb_001430_020 |
0.1330395039 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_004375_050 |
0.1335355569 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_027445_100 |
0.1343007781 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 16 |
Hb_005235_070 |
0.1348068168 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000555_020 |
0.1424780299 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000796_160 |
0.142495557 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005276_010 |
0.1429423247 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 20 |
Hb_000941_100 |
0.1430459698 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |