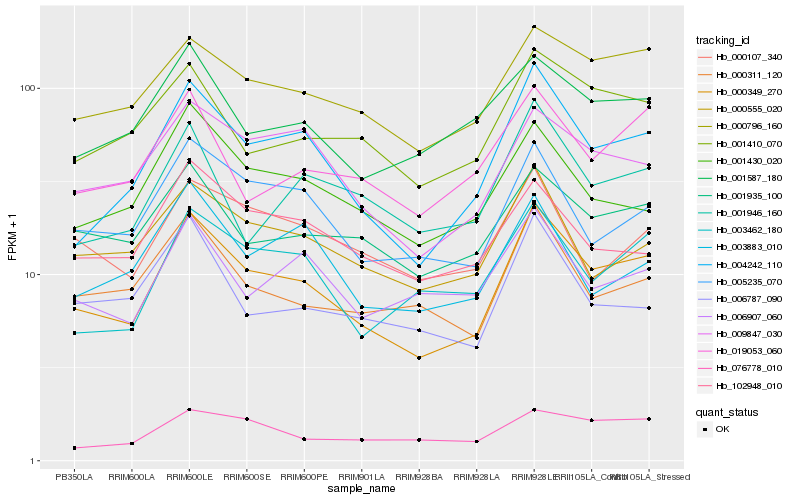
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000349_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein ycf49 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004242_110 |
0.0857855289 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 3 |
Hb_000796_160 |
0.1035921132 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005235_070 |
0.1105185634 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_009847_030 |
0.1105497546 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_001935_100 |
0.1148984791 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 7 |
Hb_001410_070 |
0.1161665581 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 8 |
Hb_000311_120 |
0.1177909868 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 9 |
Hb_003462_180 |
0.11914913 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 10 |
Hb_000555_020 |
0.1208638977 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001430_020 |
0.1225714805 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_001587_180 |
0.1253670077 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 13 |
Hb_019053_060 |
0.1265898202 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 14 |
Hb_006907_060 |
0.1293849152 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_076778_010 |
0.1297737895 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 16 |
Hb_001946_160 |
0.1304063024 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 17 |
Hb_102948_010 |
0.130974715 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 18 |
Hb_006787_090 |
0.1311000401 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 19 |
Hb_000107_340 |
0.1319730687 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 20 |
Hb_003883_010 |
0.1328210955 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |