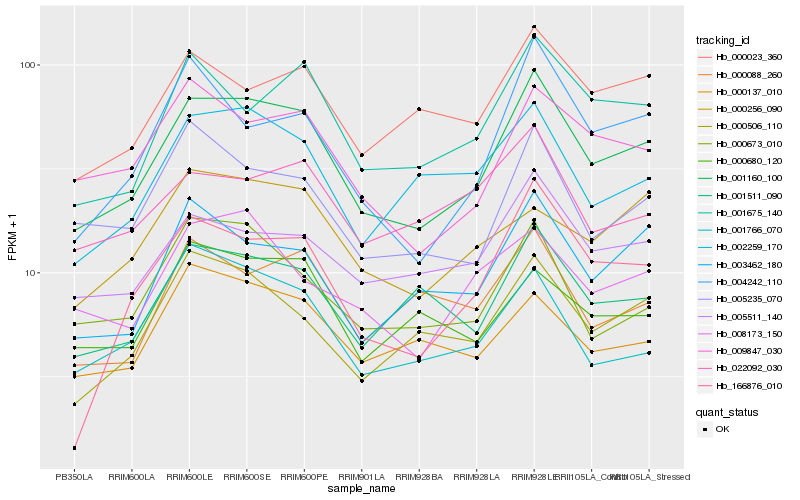
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001160_100 |
0.0 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001675_140 |
0.1034771742 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 3 |
Hb_009847_030 |
0.1048727616 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 4 |
Hb_005511_140 |
0.1058755856 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 5 |
Hb_001511_090 |
0.1065361998 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 6 |
Hb_002259_170 |
0.1078855313 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_005235_070 |
0.1081351913 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003462_180 |
0.1086621549 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 9 |
Hb_000506_110 |
0.1106663884 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 10 |
Hb_000673_010 |
0.1115980169 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000023_360 |
0.1125446046 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004242_110 |
0.1151286475 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 13 |
Hb_166876_010 |
0.1177420716 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 14 |
Hb_001766_070 |
0.1201275733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000137_010 |
0.1222826903 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 16 |
Hb_008173_150 |
0.1231721324 |
- |
- |
PREDICTED: uncharacterized protein LOC105645812 [Jatropha curcas] |
| 17 |
Hb_022092_030 |
0.123788929 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000256_090 |
0.12399393 |
- |
- |
PREDICTED: uncharacterized protein LOC105637579 [Jatropha curcas] |
| 19 |
Hb_000088_260 |
0.1241467835 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 20 |
Hb_000680_120 |
0.1241977808 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |