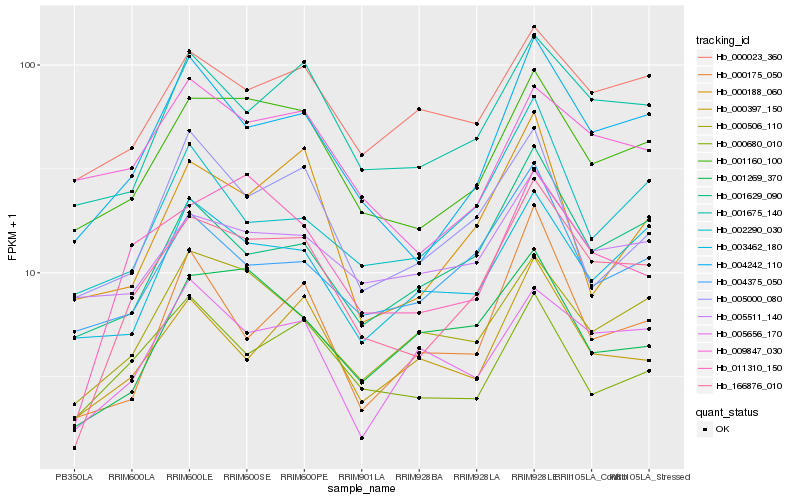
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166876_010 |
0.0 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 2 |
Hb_004242_110 |
0.1077485146 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 3 |
Hb_001160_100 |
0.1177420716 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 4 |
Hb_011310_150 |
0.1207584584 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_001675_140 |
0.1313165547 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 6 |
Hb_001629_090 |
0.1412215198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000397_150 |
0.1500866453 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 8 |
Hb_000506_110 |
0.1508534503 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 9 |
Hb_000023_360 |
0.1547205381 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000680_010 |
0.1557330424 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 11 |
Hb_004375_050 |
0.15578637 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_005656_170 |
0.1563815425 |
- |
- |
PREDICTED: uncharacterized protein LOC105637447 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003462_180 |
0.1564732955 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 14 |
Hb_001269_370 |
0.1608828579 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 15 |
Hb_005511_140 |
0.1629234786 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 16 |
Hb_000188_060 |
0.166193896 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 17 |
Hb_002290_030 |
0.1666737042 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005000_080 |
0.16706044 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 19 |
Hb_009847_030 |
0.1682553884 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_000175_050 |
0.168267345 |
- |
- |
- |