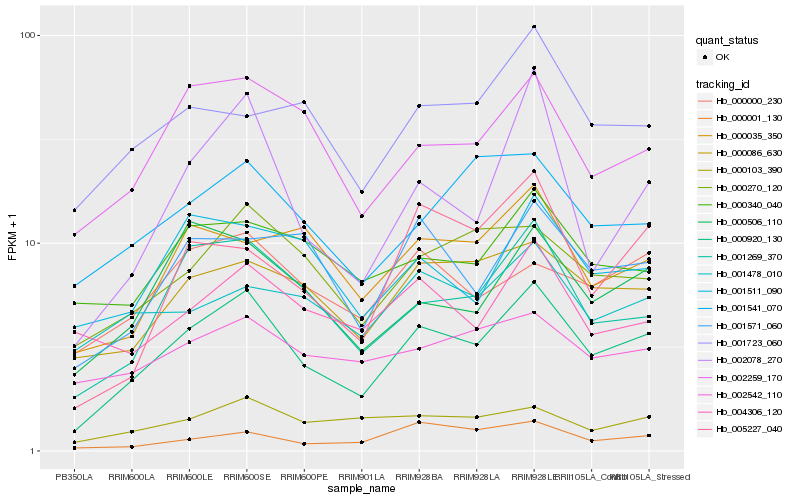
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001269_370 |
0.1188986747 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 3 |
Hb_002259_170 |
0.1345144611 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000086_630 |
0.1361262008 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000506_110 |
0.1383583041 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 6 |
Hb_000270_120 |
0.1404605223 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 7 |
Hb_001571_060 |
0.1454015805 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 8 |
Hb_000000_230 |
0.1487208008 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000103_390 |
0.1504078029 |
- |
- |
Pentatricopeptide repeat superfamily protein [Theobroma cacao] |
| 10 |
Hb_001541_070 |
0.1510468133 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 11 |
Hb_001511_090 |
0.1524616523 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 12 |
Hb_002078_270 |
0.1533836914 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 13 |
Hb_000035_350 |
0.1554065743 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001723_060 |
0.1589514456 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 15 |
Hb_005227_040 |
0.1590883164 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000340_040 |
0.1596261819 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 17 |
Hb_002542_110 |
0.1600404091 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 18 |
Hb_000001_130 |
0.1608285932 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 19 |
Hb_001478_010 |
0.1621797629 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004306_120 |
0.1624216453 |
- |
- |
DNA binding protein, putative [Ricinus communis] |