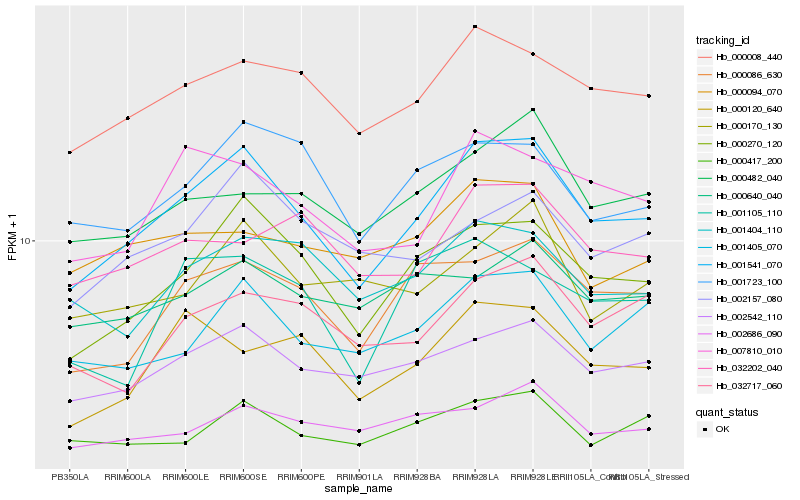
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_070 |
0.0 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 2 |
Hb_000270_120 |
0.0769570972 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 3 |
Hb_002542_110 |
0.0944050972 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 4 |
Hb_000008_440 |
0.0976880915 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |
| 5 |
Hb_001405_070 |
0.1034457923 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 6 |
Hb_000086_630 |
0.1044947665 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007810_010 |
0.108871241 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000417_200 |
0.1138452054 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002686_090 |
0.1152161132 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 10 |
Hb_001723_100 |
0.1162638437 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001404_110 |
0.1177723389 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 12 |
Hb_000482_040 |
0.1188299647 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 13 |
Hb_032717_060 |
0.1188556298 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000170_130 |
0.1212961917 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 15 |
Hb_002157_080 |
0.1215123961 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000120_640 |
0.1245820689 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_032202_040 |
0.1261724822 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 18 |
Hb_000640_040 |
0.1277020535 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000094_070 |
0.1282586316 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001105_110 |
0.1291784739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |