| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
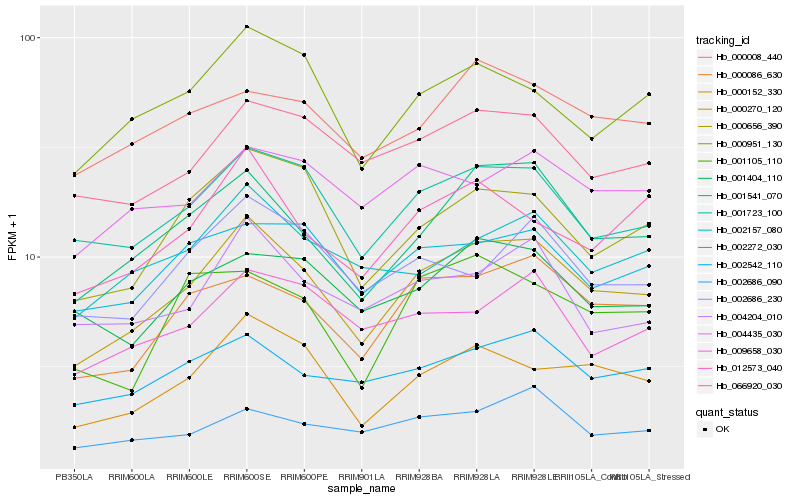
Hb_000270_120 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 2 |
Hb_001541_070 |
0.0769570972 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 3 |
Hb_000152_330 |
0.0870822163 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 4 |
Hb_001723_100 |
0.0871934365 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000086_630 |
0.088048706 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000656_390 |
0.0969629548 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002542_110 |
0.0993985291 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 8 |
Hb_000951_130 |
0.1018228005 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 9 |
Hb_012573_040 |
0.1034279543 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 10 |
Hb_066920_030 |
0.1040235562 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 11 |
Hb_002157_080 |
0.1041182695 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002272_030 |
0.1089091713 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 13 |
Hb_004204_010 |
0.108957317 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 14 |
Hb_000008_440 |
0.1118361829 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |
| 15 |
Hb_002686_230 |
0.1126197867 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_009658_030 |
0.1133309266 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 17 |
Hb_001404_110 |
0.1146532231 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 18 |
Hb_002686_090 |
0.115191447 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 19 |
Hb_001105_110 |
0.1156839022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004435_030 |
0.1159182562 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |