| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
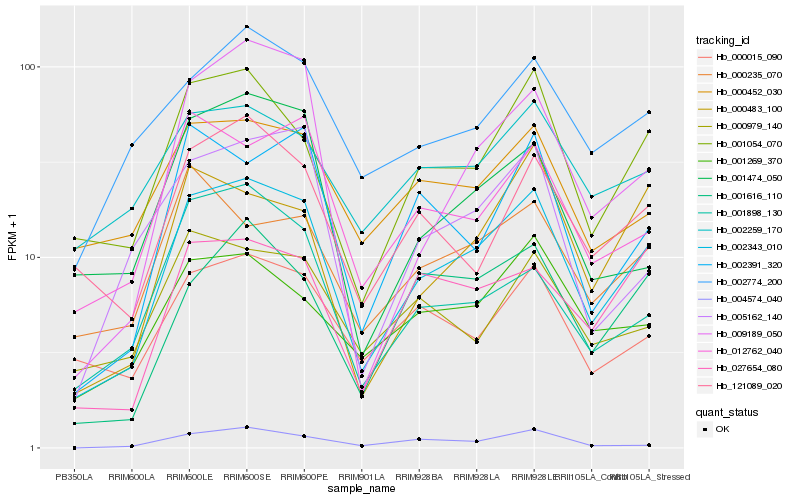
Hb_002343_010 |
0.0 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_001054_070 |
0.1199316569 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 3 |
Hb_002774_200 |
0.1421063916 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 4 |
Hb_012762_040 |
0.1456581561 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 5 |
Hb_001616_110 |
0.1458360586 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 6 |
Hb_001269_370 |
0.1483145272 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 7 |
Hb_121089_020 |
0.1484046935 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 8 |
Hb_027654_080 |
0.1486040805 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000483_100 |
0.1493890729 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 10 |
Hb_009189_050 |
0.1496315205 |
- |
- |
PREDICTED: uncharacterized protein LOC105648080 [Jatropha curcas] |
| 11 |
Hb_002391_320 |
0.1520209387 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 12 |
Hb_000452_030 |
0.1524452412 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 13 |
Hb_002259_170 |
0.1539826084 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000015_090 |
0.15475764 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 15 |
Hb_005162_140 |
0.1548502691 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 16 |
Hb_004574_040 |
0.1554597002 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 17 |
Hb_000235_070 |
0.1590989035 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 18 |
Hb_001474_050 |
0.1597872325 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000979_140 |
0.1610660187 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 20 |
Hb_001898_130 |
0.1617774783 |
- |
- |
ATP binding protein, putative [Ricinus communis] |