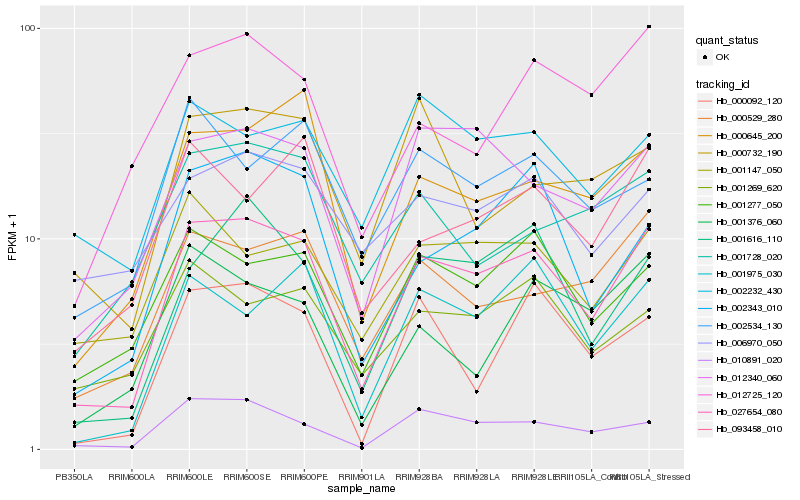
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027654_080 |
0.0 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_010891_020 |
0.1293439602 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 3 |
Hb_012340_060 |
0.1325765225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000529_280 |
0.1358160721 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001616_110 |
0.1360574892 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 6 |
Hb_001277_050 |
0.1368590768 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000645_200 |
0.1404575346 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 8 |
Hb_001975_030 |
0.1458162119 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 9 |
Hb_002343_010 |
0.1486040805 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001147_050 |
0.1500093472 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 11 |
Hb_093458_010 |
0.1540278255 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 12 |
Hb_012725_120 |
0.1564024016 |
- |
- |
PREDICTED: stress enhanced protein 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001269_620 |
0.1579956386 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 14 |
Hb_002232_430 |
0.1588327187 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000092_120 |
0.1602529518 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 16 |
Hb_001728_020 |
0.1606988559 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 17 |
Hb_002534_130 |
0.1629814225 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 18 |
Hb_006970_050 |
0.1655237592 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 19 |
Hb_000732_190 |
0.1661874624 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 20 |
Hb_001376_060 |
0.1665532182 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC isoform X2 [Jatropha curcas] |