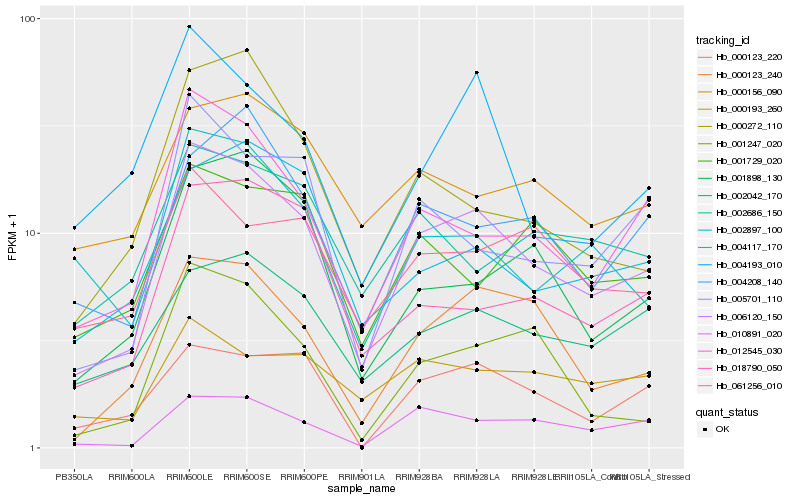
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006120_150 |
0.0 |
- |
- |
invertase [Hevea brasiliensis] |
| 2 |
Hb_012545_030 |
0.1221477463 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 3 |
Hb_000123_240 |
0.1305873323 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 4 |
Hb_010891_020 |
0.1444441908 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 5 |
Hb_005701_110 |
0.146947646 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 6 |
Hb_002042_170 |
0.1502471577 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 7 |
Hb_004208_140 |
0.1534063336 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 8 |
Hb_018790_050 |
0.1567128078 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 9 |
Hb_000123_220 |
0.1571309168 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 10 |
Hb_002897_100 |
0.1571758165 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_001898_130 |
0.1586413398 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_004117_170 |
0.1634010586 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 13 |
Hb_061256_010 |
0.1635238574 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000193_260 |
0.1650604753 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 15 |
Hb_002686_150 |
0.1653449448 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_001729_020 |
0.1655731954 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_001247_020 |
0.1661309719 |
- |
- |
galactinol synthase family protein [Populus trichocarpa] |
| 18 |
Hb_000156_090 |
0.1672223623 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000272_110 |
0.1674962601 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004193_010 |
0.1680504756 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |