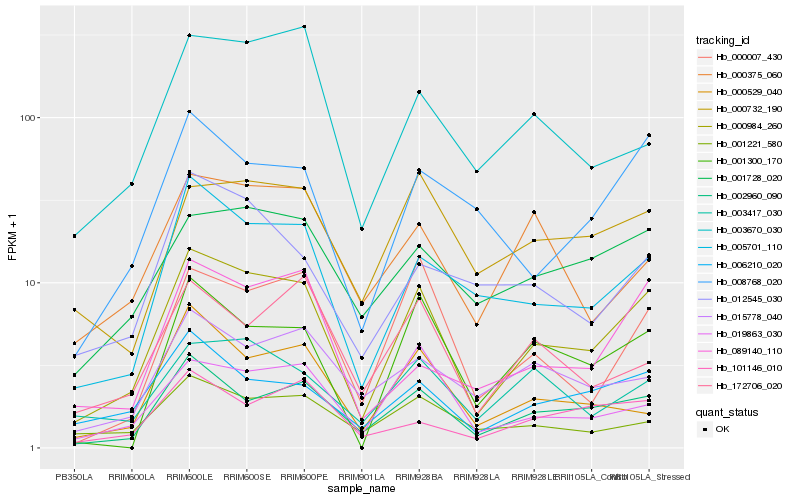
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000984_260 |
0.0 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 2 |
Hb_002960_090 |
0.1346945056 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_005701_110 |
0.1453467494 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 4 |
Hb_000007_430 |
0.1625915269 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 5 |
Hb_006210_020 |
0.1654980919 |
- |
- |
PREDICTED: calcium sensing receptor, chloroplastic [Jatropha curcas] |
| 6 |
Hb_019863_030 |
0.1674642599 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 7 |
Hb_001728_020 |
0.1721994847 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 8 |
Hb_001300_170 |
0.1747109339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000732_190 |
0.1756488053 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 10 |
Hb_008768_020 |
0.1760181216 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 11 |
Hb_003417_030 |
0.1796804844 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001221_580 |
0.1797746077 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 13 |
Hb_089140_110 |
0.1810564834 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000529_040 |
0.1833616698 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 15 |
Hb_015778_040 |
0.1868689115 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 16 |
Hb_101146_010 |
0.1894663855 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 17 |
Hb_003670_030 |
0.1896219236 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 18 |
Hb_012545_030 |
0.1912810516 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 19 |
Hb_000375_060 |
0.1912829229 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 20 |
Hb_172706_020 |
0.1916166962 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |