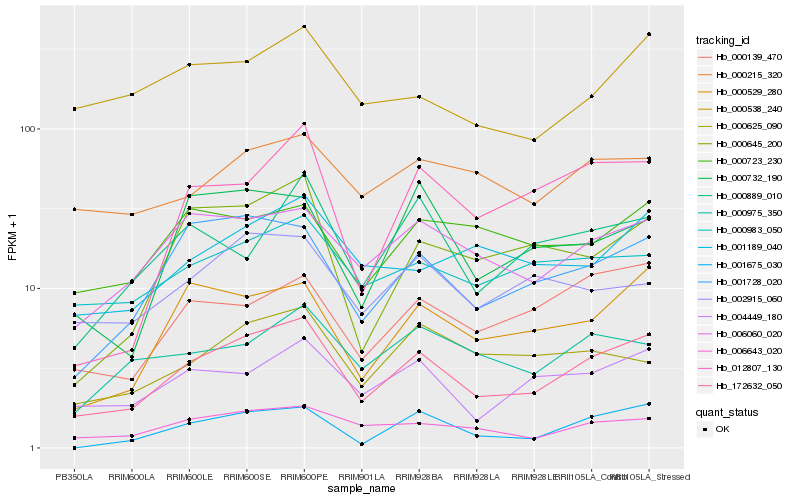
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172632_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 2 |
Hb_001728_020 |
0.1229251381 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 3 |
Hb_006060_020 |
0.1291319818 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 4 |
Hb_006643_020 |
0.1301521429 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 5 |
Hb_000529_280 |
0.131640277 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004449_180 |
0.1316623448 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 7 |
Hb_000139_470 |
0.1410529495 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_000983_050 |
0.1465209776 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_001675_030 |
0.1489881448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000645_200 |
0.1506831549 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 11 |
Hb_001189_040 |
0.1528417721 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 12 |
Hb_002915_060 |
0.1550843148 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 13 |
Hb_012807_130 |
0.1573710293 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 14 |
Hb_000625_090 |
0.1582349071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000889_010 |
0.1585576541 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 16 |
Hb_000538_240 |
0.1585803085 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 17 |
Hb_000215_320 |
0.1585860605 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 18 |
Hb_000975_350 |
0.1591131206 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 19 |
Hb_000723_230 |
0.1599267349 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 20 |
Hb_000732_190 |
0.1609155205 |
- |
- |
structural molecule, putative [Ricinus communis] |