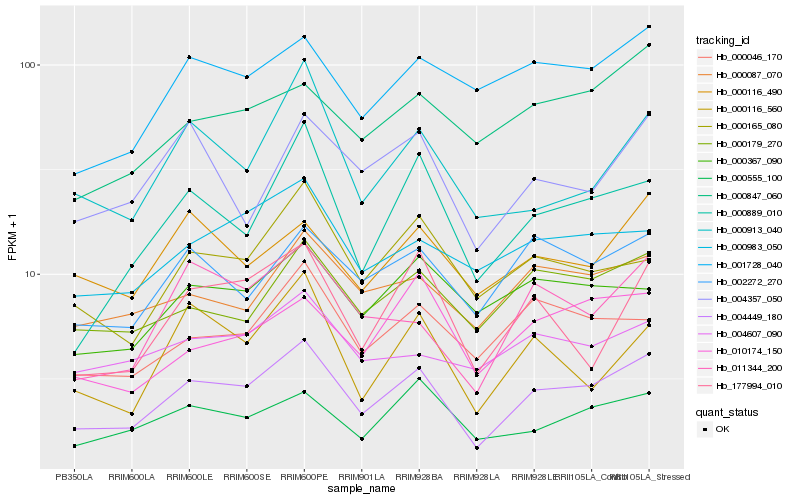
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004449_180 |
0.0 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 2 |
Hb_000179_270 |
0.1123178301 |
- |
- |
hypothetical protein JCGZ_07444 [Jatropha curcas] |
| 3 |
Hb_002272_270 |
0.1132890794 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 4 |
Hb_011344_200 |
0.1135196106 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 5 |
Hb_000555_100 |
0.1149922879 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 6 |
Hb_000889_010 |
0.1171969475 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 7 |
Hb_000087_070 |
0.1183551077 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 8 |
Hb_000046_170 |
0.1205757987 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000165_080 |
0.1222724076 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001728_040 |
0.1226587827 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 11 |
Hb_177994_010 |
0.1226645855 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 12 |
Hb_000983_050 |
0.1241345664 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000116_490 |
0.124950669 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 14 |
Hb_010174_150 |
0.1251679358 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_000847_060 |
0.1253734353 |
- |
- |
hypothetical protein [Jatropha curcas] |
| 16 |
Hb_000367_090 |
0.1258531054 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 17 |
Hb_004357_050 |
0.1270242079 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 18 |
Hb_000913_040 |
0.1279995965 |
- |
- |
Methyltransferase-related protein, putative [Theobroma cacao] |
| 19 |
Hb_000116_560 |
0.1288971823 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 20 |
Hb_004607_090 |
0.1310356327 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |