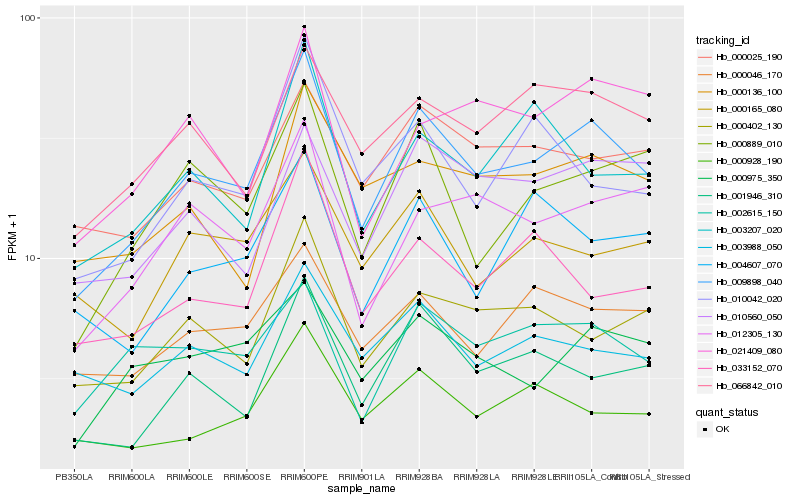
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009898_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001946_310 |
0.1113539147 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 3 |
Hb_000402_130 |
0.1155612148 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 4 |
Hb_000889_010 |
0.1163163288 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 5 |
Hb_066842_010 |
0.1166053824 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000025_190 |
0.1188700057 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 7 |
Hb_000975_350 |
0.1196016898 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 8 |
Hb_012305_130 |
0.1198295595 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 9 |
Hb_000046_170 |
0.1217238381 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000928_190 |
0.1231854396 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 11 |
Hb_010560_050 |
0.1245799561 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 12 |
Hb_002615_150 |
0.1248963749 |
- |
- |
PREDICTED: crt homolog 1-like [Jatropha curcas] |
| 13 |
Hb_000136_100 |
0.1260602892 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 14 |
Hb_003988_050 |
0.1266927036 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 15 |
Hb_021409_080 |
0.1273417568 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 16 |
Hb_003207_020 |
0.1278523912 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 17 |
Hb_000165_080 |
0.1278898366 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_033152_070 |
0.129022503 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 19 |
Hb_010042_020 |
0.1291196614 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 20 |
Hb_004607_070 |
0.1294403607 |
- |
- |
integral membrane protein, putative [Ricinus communis] |