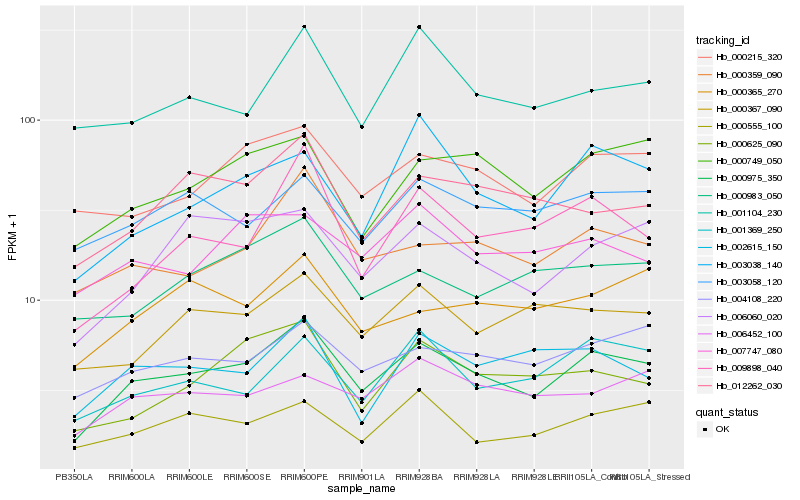
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_350 |
0.0 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 2 |
Hb_000215_320 |
0.1130094077 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 3 |
Hb_004108_220 |
0.1135412056 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000359_090 |
0.1147584734 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 5 |
Hb_006060_020 |
0.1153973625 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000749_050 |
0.1172152715 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 7 |
Hb_009898_040 |
0.1196016898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001369_250 |
0.1198064289 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012262_030 |
0.1205799567 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000367_090 |
0.1210881679 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 11 |
Hb_000625_090 |
0.1244804669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000983_050 |
0.1254460814 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_003058_120 |
0.1258386778 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 14 |
Hb_001104_230 |
0.1261333163 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 15 |
Hb_000555_100 |
0.1261884095 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 16 |
Hb_003038_140 |
0.1275729499 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 17 |
Hb_002615_150 |
0.1278577003 |
- |
- |
PREDICTED: crt homolog 1-like [Jatropha curcas] |
| 18 |
Hb_006452_100 |
0.1279251564 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 19 |
Hb_000365_270 |
0.1285004118 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 20 |
Hb_007747_080 |
0.1301026257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |