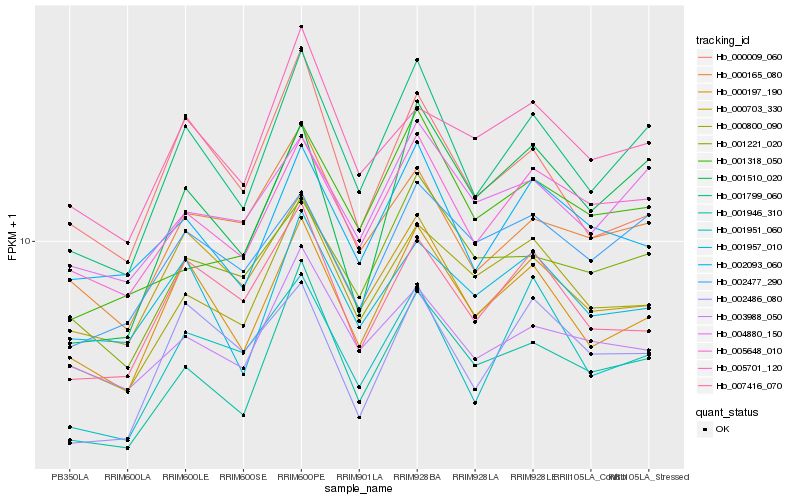
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001799_060 |
0.0 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 2 |
Hb_000197_190 |
0.0749391644 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001946_310 |
0.0841321137 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 4 |
Hb_005648_010 |
0.0866055351 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 5 |
Hb_000800_090 |
0.0999897744 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 6 |
Hb_004880_150 |
0.1017981004 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 7 |
Hb_000165_080 |
0.1026438405 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002093_060 |
0.1037790113 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_001951_060 |
0.1060573548 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 10 |
Hb_007416_070 |
0.1064182803 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 11 |
Hb_001221_020 |
0.1072094056 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 12 |
Hb_000703_330 |
0.1074256623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001510_020 |
0.1081596065 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 14 |
Hb_002477_290 |
0.109564339 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_005701_120 |
0.1139419478 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 16 |
Hb_001957_010 |
0.1146786107 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003988_050 |
0.116049727 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 18 |
Hb_001318_050 |
0.1169036526 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_000009_060 |
0.1175599054 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 20 |
Hb_002486_080 |
0.1182577196 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |