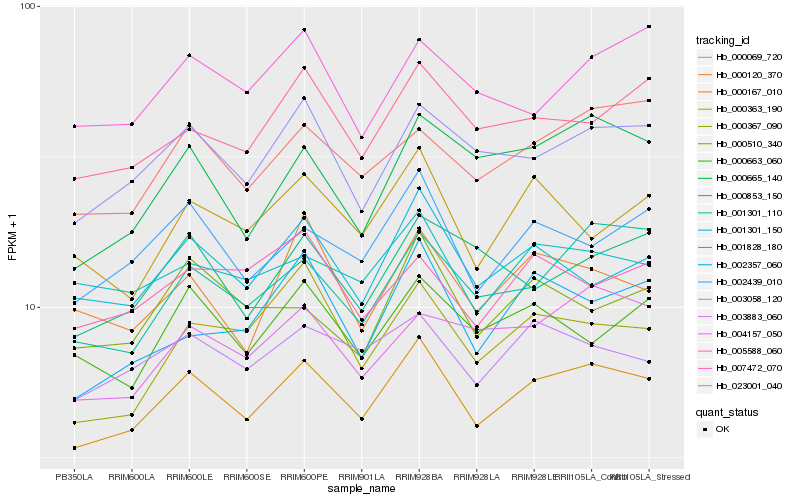
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000167_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003058_120 |
0.060855721 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 3 |
Hb_002439_010 |
0.0627371749 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 4 |
Hb_000069_720 |
0.0676460593 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 5 |
Hb_003883_060 |
0.068488382 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001301_150 |
0.0702234171 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 7 |
Hb_023001_040 |
0.0703917756 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_001301_110 |
0.0711248847 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 9 |
Hb_001828_180 |
0.0711624151 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 10 |
Hb_004157_050 |
0.0770699726 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000665_140 |
0.078017992 |
- |
- |
PREDICTED: protein MEMO1 [Jatropha curcas] |
| 12 |
Hb_000663_060 |
0.0787258267 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 13 |
Hb_000363_190 |
0.079434865 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000120_370 |
0.0798813937 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 15 |
Hb_000367_090 |
0.0811483512 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 16 |
Hb_000510_340 |
0.0815447304 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 17 |
Hb_002357_060 |
0.0815975532 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 18 |
Hb_007472_070 |
0.0819970181 |
- |
- |
cir, putative [Ricinus communis] |
| 19 |
Hb_000853_150 |
0.0822045455 |
- |
- |
Fumarase 1 isoform 2 [Theobroma cacao] |
| 20 |
Hb_005588_060 |
0.0835843456 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |