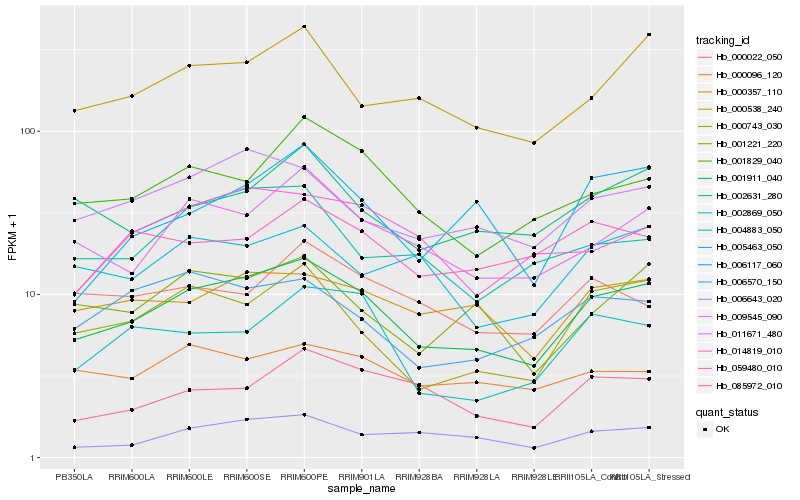
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001911_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 2 |
Hb_011671_480 |
0.1065220847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009545_090 |
0.1129804849 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 4 |
Hb_000538_240 |
0.1133519278 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 5 |
Hb_000743_030 |
0.1150347881 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 6 |
Hb_001829_040 |
0.116789833 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 7 |
Hb_000357_110 |
0.1179837542 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 8 |
Hb_006643_020 |
0.1186469288 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 9 |
Hb_002631_280 |
0.1210720417 |
- |
- |
PREDICTED: ubiquitin isoform X2 [Eucalyptus grandis] |
| 10 |
Hb_001221_220 |
0.1232596004 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 11 |
Hb_085972_010 |
0.1252630865 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 12 |
Hb_002869_050 |
0.12880289 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_004883_050 |
0.1296539708 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 14 |
Hb_006570_150 |
0.1303574587 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005463_050 |
0.1306476934 |
- |
- |
PREDICTED: uncharacterized protein LOC105632514 [Jatropha curcas] |
| 16 |
Hb_014819_010 |
0.1310711313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000096_120 |
0.1319416323 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 18 |
Hb_059480_010 |
0.1322006661 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 19 |
Hb_006117_060 |
0.1358488854 |
- |
- |
hypothetical protein CARUB_v10024452mg [Capsella rubella] |
| 20 |
Hb_000022_050 |
0.1376027763 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |