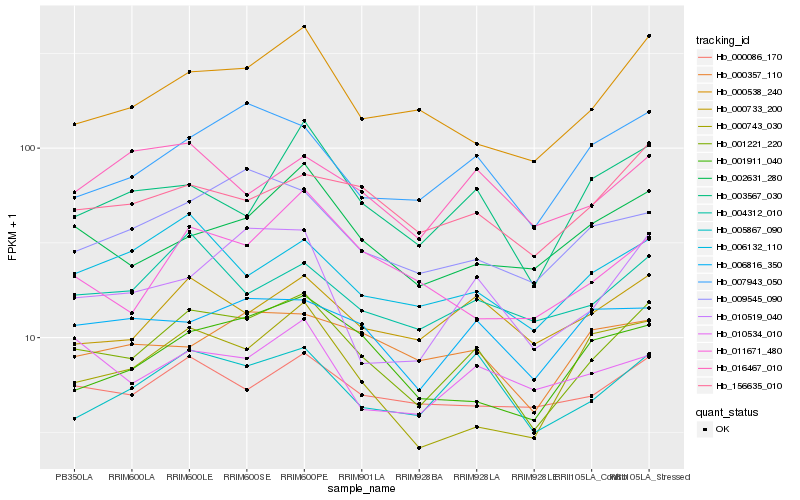
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_220 |
0.0 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 2 |
Hb_005867_090 |
0.1080949359 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 3 |
Hb_000538_240 |
0.1126627589 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 4 |
Hb_007943_050 |
0.1167681898 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 5 |
Hb_156635_010 |
0.1186503152 |
- |
- |
PREDICTED: autophagy-related protein 8f [Jatropha curcas] |
| 6 |
Hb_000733_200 |
0.1202591071 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 7 |
Hb_003567_030 |
0.1212099442 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |
| 8 |
Hb_010519_040 |
0.1224434254 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 9 |
Hb_006132_110 |
0.1228077469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004312_010 |
0.1228584661 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001911_040 |
0.1232596004 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 12 |
Hb_002631_280 |
0.1236371784 |
- |
- |
PREDICTED: ubiquitin isoform X2 [Eucalyptus grandis] |
| 13 |
Hb_009545_090 |
0.1240563599 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 14 |
Hb_006816_350 |
0.1241960708 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |
| 15 |
Hb_000086_170 |
0.1242429815 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 16 |
Hb_000357_110 |
0.1324663322 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000743_030 |
0.1330419477 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 18 |
Hb_011671_480 |
0.1337241377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010534_010 |
0.1359326692 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 20 |
Hb_016467_010 |
0.1374706644 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |