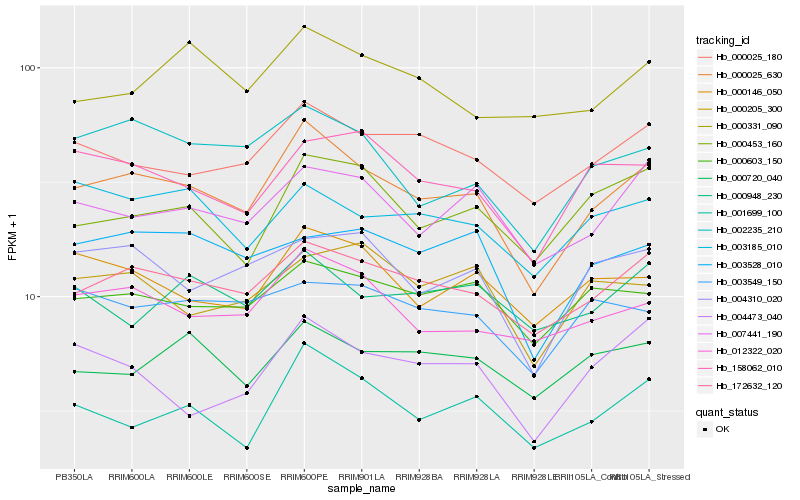
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_630 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635952 [Jatropha curcas] |
| 2 |
Hb_172632_120 |
0.068249979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012322_020 |
0.0869795787 |
- |
- |
- |
| 4 |
Hb_000025_180 |
0.0882655834 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 5 |
Hb_002235_210 |
0.0895102077 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 6 |
Hb_001699_100 |
0.0909684755 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 7 |
Hb_007441_190 |
0.0919944788 |
- |
- |
hypothetical protein CISIN_1g023046mg [Citrus sinensis] |
| 8 |
Hb_000146_050 |
0.0933973391 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 9 |
Hb_004310_020 |
0.0954035331 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_158062_010 |
0.0957086954 |
- |
- |
PREDICTED: uncharacterized protein LOC105634413 [Jatropha curcas] |
| 11 |
Hb_000603_150 |
0.0965335454 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 12 |
Hb_000453_160 |
0.0965832177 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 13 |
Hb_000720_040 |
0.1034248901 |
- |
- |
PREDICTED: replication factor C subunit 2 [Jatropha curcas] |
| 14 |
Hb_003185_010 |
0.1037074529 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |
| 15 |
Hb_003528_010 |
0.1042457752 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 3R-1 [Jatropha curcas] |
| 16 |
Hb_004473_040 |
0.1053460931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000205_300 |
0.1065821946 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 18 |
Hb_003549_150 |
0.1067599455 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 19 |
Hb_000331_090 |
0.1083944485 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 20 |
Hb_000948_230 |
0.1086665932 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |