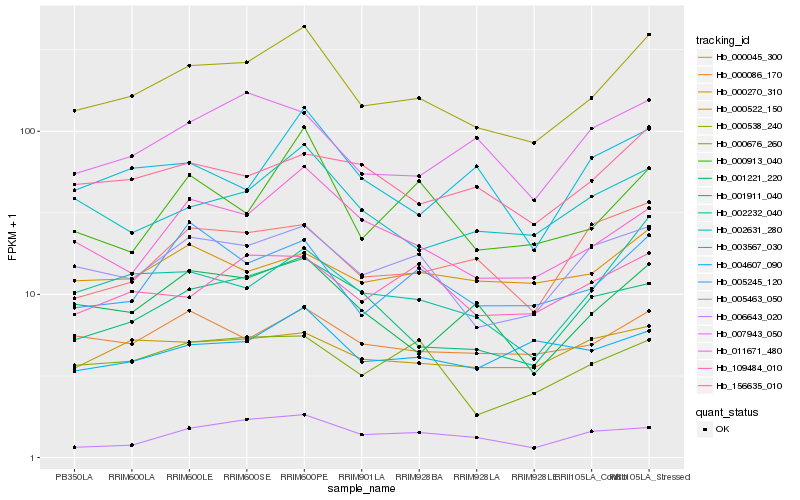
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 2 |
Hb_005463_050 |
0.1022396937 |
- |
- |
PREDICTED: uncharacterized protein LOC105632514 [Jatropha curcas] |
| 3 |
Hb_011671_480 |
0.1057915267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002232_040 |
0.1078887881 |
- |
- |
- |
| 5 |
Hb_002631_280 |
0.1106473364 |
- |
- |
PREDICTED: ubiquitin isoform X2 [Eucalyptus grandis] |
| 6 |
Hb_001221_220 |
0.1126627589 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 7 |
Hb_001911_040 |
0.1133519278 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 8 |
Hb_000086_170 |
0.1181838947 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 9 |
Hb_005245_120 |
0.1209788733 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 10 |
Hb_156635_010 |
0.125519107 |
- |
- |
PREDICTED: autophagy-related protein 8f [Jatropha curcas] |
| 11 |
Hb_109484_010 |
0.1267164857 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 12 |
Hb_000045_300 |
0.1273852708 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 13 |
Hb_000913_040 |
0.1291216068 |
- |
- |
Methyltransferase-related protein, putative [Theobroma cacao] |
| 14 |
Hb_000676_260 |
0.1299733662 |
- |
- |
PREDICTED: uncharacterized protein LOC105639070 [Jatropha curcas] |
| 15 |
Hb_004607_090 |
0.1305539155 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 16 |
Hb_000522_150 |
0.1313612474 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 17 |
Hb_007943_050 |
0.1316756871 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 18 |
Hb_006643_020 |
0.1318315704 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 19 |
Hb_003567_030 |
0.1324083686 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000270_310 |
0.1332298056 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |